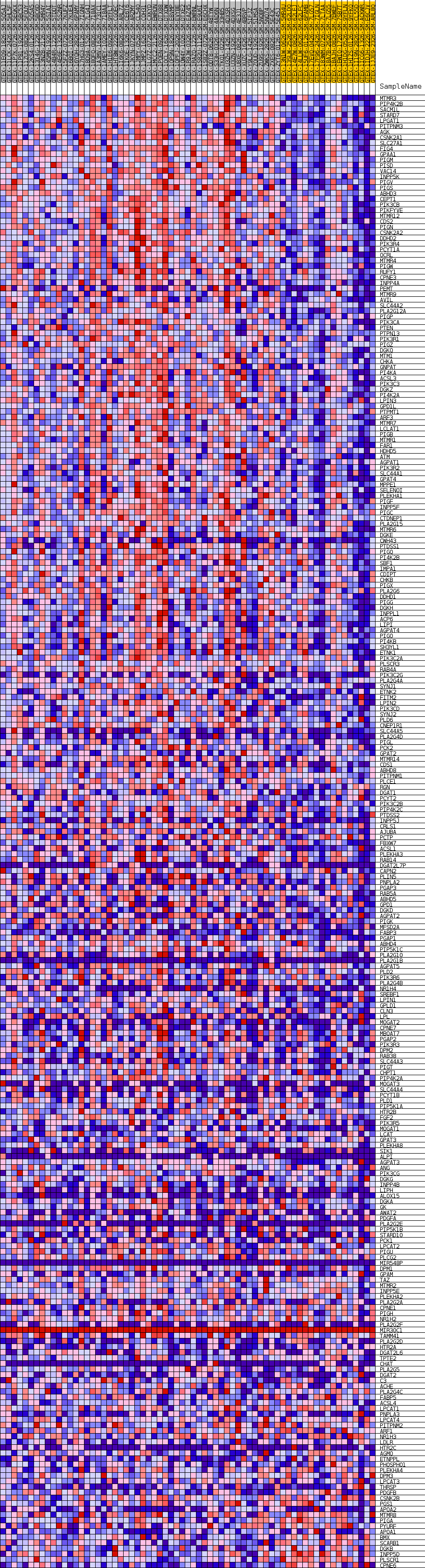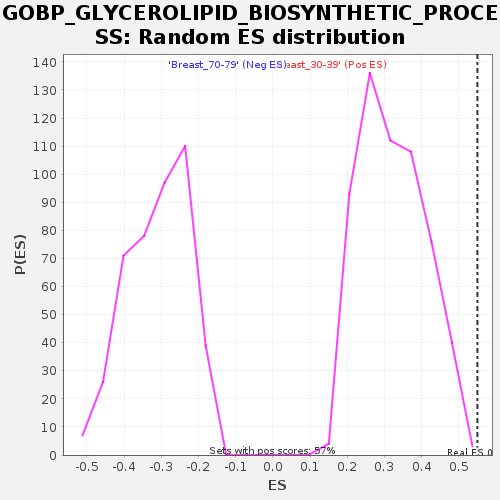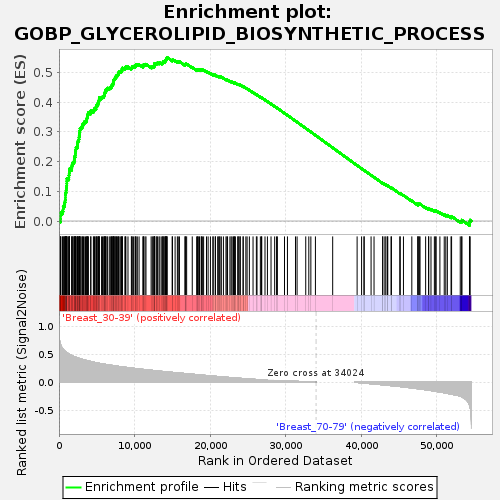
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Breast.Breast.cls #Breast_30-39_versus_Breast_70-79.Breast.cls #Breast_30-39_versus_Breast_70-79_repos |
| Phenotype | Breast.cls#Breast_30-39_versus_Breast_70-79_repos |
| Upregulated in class | Breast_30-39 |
| GeneSet | GOBP_GLYCEROLIPID_BIOSYNTHETIC_PROCESS |
| Enrichment Score (ES) | 0.55003476 |
| Normalized Enrichment Score (NES) | 1.7017386 |
| Nominal p-value | 0.0017482517 |
| FDR q-value | 0.336399 |
| FWER p-Value | 0.645 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | MTMR3 | NA | 209 | 0.668 | 0.0067 | Yes |
| 2 | PIP4K2B | NA | 221 | 0.663 | 0.0170 | Yes |
| 3 | SACM1L | NA | 227 | 0.660 | 0.0273 | Yes |
| 4 | STARD7 | NA | 457 | 0.603 | 0.0326 | Yes |
| 5 | LPGAT1 | NA | 546 | 0.586 | 0.0402 | Yes |
| 6 | PITPNM3 | NA | 558 | 0.584 | 0.0492 | Yes |
| 7 | AGK | NA | 698 | 0.562 | 0.0555 | Yes |
| 8 | CSNK2A1 | NA | 759 | 0.556 | 0.0632 | Yes |
| 9 | SLC27A1 | NA | 819 | 0.549 | 0.0708 | Yes |
| 10 | FIG4 | NA | 851 | 0.545 | 0.0788 | Yes |
| 11 | GPAA1 | NA | 854 | 0.545 | 0.0874 | Yes |
| 12 | PIGM | NA | 877 | 0.543 | 0.0956 | Yes |
| 13 | PISD | NA | 944 | 0.536 | 0.1028 | Yes |
| 14 | VAC14 | NA | 970 | 0.533 | 0.1107 | Yes |
| 15 | INPP5K | NA | 1017 | 0.528 | 0.1182 | Yes |
| 16 | PIGV | NA | 1029 | 0.528 | 0.1264 | Yes |
| 17 | PIGS | NA | 1039 | 0.526 | 0.1345 | Yes |
| 18 | ABHD3 | NA | 1049 | 0.525 | 0.1426 | Yes |
| 19 | CEPT1 | NA | 1300 | 0.503 | 0.1460 | Yes |
| 20 | PIK3CB | NA | 1307 | 0.502 | 0.1538 | Yes |
| 21 | PIKFYVE | NA | 1353 | 0.499 | 0.1608 | Yes |
| 22 | MTMR12 | NA | 1368 | 0.497 | 0.1684 | Yes |
| 23 | CDS2 | NA | 1391 | 0.496 | 0.1758 | Yes |
| 24 | PIGN | NA | 1685 | 0.477 | 0.1780 | Yes |
| 25 | CSNK2A2 | NA | 1703 | 0.476 | 0.1852 | Yes |
| 26 | DDHD2 | NA | 1743 | 0.473 | 0.1919 | Yes |
| 27 | PIK3R4 | NA | 1840 | 0.466 | 0.1975 | Yes |
| 28 | PCYT1A | NA | 1993 | 0.456 | 0.2019 | Yes |
| 29 | OCRL | NA | 2023 | 0.454 | 0.2085 | Yes |
| 30 | MTMR4 | NA | 2037 | 0.454 | 0.2154 | Yes |
| 31 | PIGW | NA | 2130 | 0.449 | 0.2208 | Yes |
| 32 | RUFY1 | NA | 2161 | 0.448 | 0.2273 | Yes |
| 33 | CPNE3 | NA | 2172 | 0.448 | 0.2342 | Yes |
| 34 | INPP4A | NA | 2217 | 0.445 | 0.2404 | Yes |
| 35 | PEMT | NA | 2261 | 0.442 | 0.2466 | Yes |
| 36 | MTMR9 | NA | 2410 | 0.435 | 0.2508 | Yes |
| 37 | AVIL | NA | 2444 | 0.433 | 0.2570 | Yes |
| 38 | SLC44A2 | NA | 2472 | 0.432 | 0.2633 | Yes |
| 39 | PLA2G12A | NA | 2515 | 0.430 | 0.2693 | Yes |
| 40 | PIGP | NA | 2621 | 0.426 | 0.2741 | Yes |
| 41 | PIK3CA | NA | 2652 | 0.424 | 0.2803 | Yes |
| 42 | PTEN | NA | 2673 | 0.423 | 0.2866 | Yes |
| 43 | PTPN13 | NA | 2705 | 0.422 | 0.2927 | Yes |
| 44 | PIK3R1 | NA | 2739 | 0.420 | 0.2987 | Yes |
| 45 | PIGZ | NA | 2762 | 0.419 | 0.3049 | Yes |
| 46 | DGKQ | NA | 2788 | 0.418 | 0.3110 | Yes |
| 47 | MTM1 | NA | 2916 | 0.412 | 0.3152 | Yes |
| 48 | CHKA | NA | 3068 | 0.406 | 0.3188 | Yes |
| 49 | GNPAT | NA | 3103 | 0.405 | 0.3246 | Yes |
| 50 | PI4KA | NA | 3246 | 0.400 | 0.3283 | Yes |
| 51 | ACSL3 | NA | 3331 | 0.397 | 0.3330 | Yes |
| 52 | PIK3C3 | NA | 3507 | 0.391 | 0.3360 | Yes |
| 53 | DGKZ | NA | 3668 | 0.385 | 0.3391 | Yes |
| 54 | PI4K2A | NA | 3675 | 0.385 | 0.3451 | Yes |
| 55 | LPIN3 | NA | 3734 | 0.382 | 0.3500 | Yes |
| 56 | GPD1L | NA | 3756 | 0.382 | 0.3557 | Yes |
| 57 | PTPMT1 | NA | 3833 | 0.379 | 0.3602 | Yes |
| 58 | ARF3 | NA | 3888 | 0.377 | 0.3652 | Yes |
| 59 | MTMR7 | NA | 4199 | 0.368 | 0.3653 | Yes |
| 60 | LCLAT1 | NA | 4217 | 0.368 | 0.3708 | Yes |
| 61 | PIGB | NA | 4552 | 0.358 | 0.3703 | Yes |
| 62 | MTMR1 | NA | 4630 | 0.356 | 0.3745 | Yes |
| 63 | FAR1 | NA | 4703 | 0.353 | 0.3787 | Yes |
| 64 | HDHD5 | NA | 4894 | 0.348 | 0.3807 | Yes |
| 65 | ATM | NA | 4915 | 0.348 | 0.3858 | Yes |
| 66 | AGPAT1 | NA | 4979 | 0.346 | 0.3901 | Yes |
| 67 | PIK3R2 | NA | 5143 | 0.342 | 0.3925 | Yes |
| 68 | SLC44A1 | NA | 5167 | 0.341 | 0.3975 | Yes |
| 69 | GPAT4 | NA | 5223 | 0.339 | 0.4018 | Yes |
| 70 | MPPE1 | NA | 5286 | 0.338 | 0.4060 | Yes |
| 71 | SELENOI | NA | 5319 | 0.337 | 0.4107 | Yes |
| 72 | PLEKHA1 | NA | 5339 | 0.336 | 0.4157 | Yes |
| 73 | PIGF | NA | 5633 | 0.329 | 0.4155 | Yes |
| 74 | INPP5F | NA | 5763 | 0.326 | 0.4183 | Yes |
| 75 | PIGC | NA | 5925 | 0.322 | 0.4204 | Yes |
| 76 | CTDNEP1 | NA | 5991 | 0.321 | 0.4242 | Yes |
| 77 | PLA2G15 | NA | 6047 | 0.320 | 0.4283 | Yes |
| 78 | MTMR6 | NA | 6066 | 0.319 | 0.4330 | Yes |
| 79 | DGKE | NA | 6163 | 0.317 | 0.4362 | Yes |
| 80 | CWH43 | NA | 6171 | 0.317 | 0.4411 | Yes |
| 81 | PTDSS1 | NA | 6348 | 0.313 | 0.4428 | Yes |
| 82 | PIGQ | NA | 6397 | 0.312 | 0.4468 | Yes |
| 83 | PI4K2B | NA | 6648 | 0.306 | 0.4470 | Yes |
| 84 | SBF1 | NA | 6786 | 0.303 | 0.4493 | Yes |
| 85 | IMPA1 | NA | 6915 | 0.301 | 0.4517 | Yes |
| 86 | CDIPT | NA | 6957 | 0.300 | 0.4557 | Yes |
| 87 | CHKB | NA | 7051 | 0.298 | 0.4587 | Yes |
| 88 | PIGX | NA | 7148 | 0.296 | 0.4616 | Yes |
| 89 | PLA2G6 | NA | 7179 | 0.296 | 0.4657 | Yes |
| 90 | DDHD1 | NA | 7190 | 0.295 | 0.4702 | Yes |
| 91 | PIGG | NA | 7224 | 0.295 | 0.4742 | Yes |
| 92 | DGKH | NA | 7314 | 0.293 | 0.4772 | Yes |
| 93 | INPPL1 | NA | 7421 | 0.290 | 0.4798 | Yes |
| 94 | ACP6 | NA | 7440 | 0.290 | 0.4841 | Yes |
| 95 | LIPI | NA | 7580 | 0.287 | 0.4860 | Yes |
| 96 | AGPAT4 | NA | 7620 | 0.287 | 0.4898 | Yes |
| 97 | PIGO | NA | 7764 | 0.284 | 0.4917 | Yes |
| 98 | PI4KB | NA | 7834 | 0.282 | 0.4949 | Yes |
| 99 | SH3YL1 | NA | 7864 | 0.282 | 0.4988 | Yes |
| 100 | ETNK1 | NA | 7952 | 0.280 | 0.5016 | Yes |
| 101 | PIK3C2A | NA | 8112 | 0.277 | 0.5030 | Yes |
| 102 | PLSCR3 | NA | 8268 | 0.274 | 0.5045 | Yes |
| 103 | RAB4A | NA | 8331 | 0.273 | 0.5077 | Yes |
| 104 | PIK3C2G | NA | 8342 | 0.273 | 0.5118 | Yes |
| 105 | PLA2G4A | NA | 8399 | 0.272 | 0.5151 | Yes |
| 106 | SYNJ1 | NA | 8722 | 0.266 | 0.5133 | Yes |
| 107 | ETNK2 | NA | 8802 | 0.265 | 0.5161 | Yes |
| 108 | FITM2 | NA | 8853 | 0.264 | 0.5193 | Yes |
| 109 | LPIN2 | NA | 9132 | 0.259 | 0.5183 | Yes |
| 110 | PIK3CD | NA | 9579 | 0.251 | 0.5140 | Yes |
| 111 | SYNJ2 | NA | 9642 | 0.250 | 0.5168 | Yes |
| 112 | PLD6 | NA | 9694 | 0.249 | 0.5198 | Yes |
| 113 | CNEP1R1 | NA | 9874 | 0.247 | 0.5204 | Yes |
| 114 | SLC44A5 | NA | 10080 | 0.243 | 0.5205 | Yes |
| 115 | PLA2G4D | NA | 10114 | 0.243 | 0.5237 | Yes |
| 116 | PIGL | NA | 10207 | 0.241 | 0.5258 | Yes |
| 117 | PCK2 | NA | 10364 | 0.239 | 0.5267 | Yes |
| 118 | GPAT2 | NA | 10600 | 0.235 | 0.5261 | Yes |
| 119 | MTMR14 | NA | 11106 | 0.227 | 0.5204 | Yes |
| 120 | CDS1 | NA | 11169 | 0.227 | 0.5228 | Yes |
| 121 | ABHD8 | NA | 11193 | 0.226 | 0.5260 | Yes |
| 122 | PITPNM1 | NA | 11378 | 0.223 | 0.5261 | Yes |
| 123 | PLCE1 | NA | 11513 | 0.221 | 0.5271 | Yes |
| 124 | RGN | NA | 12195 | 0.212 | 0.5179 | Yes |
| 125 | DGAT1 | NA | 12349 | 0.210 | 0.5184 | Yes |
| 126 | PCYT2 | NA | 12476 | 0.208 | 0.5194 | Yes |
| 127 | PIK3C2B | NA | 12594 | 0.206 | 0.5205 | Yes |
| 128 | PIP4K2C | NA | 12621 | 0.206 | 0.5233 | Yes |
| 129 | PTDSS2 | NA | 12630 | 0.206 | 0.5264 | Yes |
| 130 | INPP5J | NA | 12632 | 0.206 | 0.5296 | Yes |
| 131 | CRLS1 | NA | 12850 | 0.203 | 0.5288 | Yes |
| 132 | AJUBA | NA | 12946 | 0.201 | 0.5303 | Yes |
| 133 | PCTP | NA | 13027 | 0.200 | 0.5320 | Yes |
| 134 | FBXW7 | NA | 13238 | 0.197 | 0.5312 | Yes |
| 135 | ACSL1 | NA | 13362 | 0.196 | 0.5320 | Yes |
| 136 | PLEKHA3 | NA | 13606 | 0.192 | 0.5306 | Yes |
| 137 | RAB14 | NA | 13732 | 0.190 | 0.5313 | Yes |
| 138 | DGAT2L7P | NA | 13778 | 0.190 | 0.5334 | Yes |
| 139 | CAPN2 | NA | 13794 | 0.190 | 0.5362 | Yes |
| 140 | PLIN5 | NA | 14018 | 0.186 | 0.5350 | Yes |
| 141 | PNPLA2 | NA | 14029 | 0.186 | 0.5378 | Yes |
| 142 | PGAP3 | NA | 14096 | 0.185 | 0.5395 | Yes |
| 143 | RAB5A | NA | 14128 | 0.185 | 0.5418 | Yes |
| 144 | ABHD5 | NA | 14212 | 0.184 | 0.5432 | Yes |
| 145 | GPD1 | NA | 14218 | 0.184 | 0.5460 | Yes |
| 146 | DGKD | NA | 14282 | 0.183 | 0.5477 | Yes |
| 147 | AGPAT2 | NA | 14313 | 0.182 | 0.5500 | Yes |
| 148 | PIGK | NA | 14980 | 0.174 | 0.5405 | No |
| 149 | MFSD2A | NA | 15023 | 0.173 | 0.5425 | No |
| 150 | FABP3 | NA | 15369 | 0.169 | 0.5388 | No |
| 151 | PGAP1 | NA | 15667 | 0.166 | 0.5359 | No |
| 152 | ABHD4 | NA | 15785 | 0.164 | 0.5364 | No |
| 153 | PIP5K1C | NA | 15944 | 0.162 | 0.5360 | No |
| 154 | PLA2G10 | NA | 16686 | 0.153 | 0.5248 | No |
| 155 | PLA2G1B | NA | 16700 | 0.153 | 0.5270 | No |
| 156 | AGPAT5 | NA | 16776 | 0.152 | 0.5280 | No |
| 157 | PLD2 | NA | 16898 | 0.150 | 0.5281 | No |
| 158 | PIK3R6 | NA | 17640 | 0.141 | 0.5167 | No |
| 159 | PLA2G4B | NA | 18222 | 0.134 | 0.5082 | No |
| 160 | NR1H4 | NA | 18353 | 0.132 | 0.5078 | No |
| 161 | SREBF1 | NA | 18357 | 0.132 | 0.5099 | No |
| 162 | LPIN1 | NA | 18498 | 0.130 | 0.5094 | No |
| 163 | GPLD1 | NA | 18598 | 0.129 | 0.5096 | No |
| 164 | CLN3 | NA | 18819 | 0.126 | 0.5075 | No |
| 165 | LPL | NA | 18871 | 0.125 | 0.5085 | No |
| 166 | MOGAT2 | NA | 18978 | 0.124 | 0.5085 | No |
| 167 | CPNE7 | NA | 19143 | 0.122 | 0.5075 | No |
| 168 | MBOAT7 | NA | 19526 | 0.117 | 0.5023 | No |
| 169 | PGAP2 | NA | 19740 | 0.114 | 0.5002 | No |
| 170 | PIK3R3 | NA | 20047 | 0.111 | 0.4963 | No |
| 171 | DPM2 | NA | 20379 | 0.107 | 0.4919 | No |
| 172 | RAB38 | NA | 20404 | 0.106 | 0.4931 | No |
| 173 | SLC44A3 | NA | 20682 | 0.103 | 0.4896 | No |
| 174 | PIGT | NA | 20709 | 0.103 | 0.4908 | No |
| 175 | CHPT1 | NA | 21006 | 0.099 | 0.4869 | No |
| 176 | PIP4K2A | NA | 21146 | 0.098 | 0.4859 | No |
| 177 | MOGAT3 | NA | 21214 | 0.097 | 0.4862 | No |
| 178 | SLC44A4 | NA | 21317 | 0.096 | 0.4858 | No |
| 179 | PCYT1B | NA | 21489 | 0.094 | 0.4842 | No |
| 180 | PLD1 | NA | 21765 | 0.091 | 0.4805 | No |
| 181 | PIP5K1A | NA | 22104 | 0.087 | 0.4757 | No |
| 182 | HTR2B | NA | 22228 | 0.086 | 0.4748 | No |
| 183 | FGF2 | NA | 22312 | 0.085 | 0.4746 | No |
| 184 | PIK3R5 | NA | 22627 | 0.081 | 0.4701 | No |
| 185 | MOGAT1 | NA | 22786 | 0.079 | 0.4684 | No |
| 186 | LCAT | NA | 22918 | 0.078 | 0.4672 | No |
| 187 | GPAT3 | NA | 23098 | 0.076 | 0.4652 | No |
| 188 | PLEKHA8 | NA | 23200 | 0.075 | 0.4645 | No |
| 189 | SIK1 | NA | 23215 | 0.075 | 0.4654 | No |
| 190 | ALPI | NA | 23354 | 0.073 | 0.4640 | No |
| 191 | AGPAT3 | NA | 23670 | 0.070 | 0.4593 | No |
| 192 | ANG | NA | 23702 | 0.070 | 0.4599 | No |
| 193 | PIK3CG | NA | 23809 | 0.069 | 0.4590 | No |
| 194 | DGKG | NA | 23961 | 0.067 | 0.4573 | No |
| 195 | INPP4B | NA | 23997 | 0.067 | 0.4577 | No |
| 196 | LIPH | NA | 24369 | 0.063 | 0.4519 | No |
| 197 | ALOX15 | NA | 24387 | 0.063 | 0.4525 | No |
| 198 | DGKA | NA | 24737 | 0.060 | 0.4471 | No |
| 199 | GK | NA | 24899 | 0.058 | 0.4450 | No |
| 200 | AWAT2 | NA | 25201 | 0.055 | 0.4404 | No |
| 201 | PDGFA | NA | 25687 | 0.051 | 0.4322 | No |
| 202 | PLA2G2E | NA | 26126 | 0.047 | 0.4249 | No |
| 203 | PIP5K1B | NA | 26213 | 0.046 | 0.4240 | No |
| 204 | STARD10 | NA | 26655 | 0.042 | 0.4166 | No |
| 205 | PCK1 | NA | 26802 | 0.041 | 0.4145 | No |
| 206 | LPCAT2 | NA | 26841 | 0.041 | 0.4145 | No |
| 207 | PIGU | NA | 27270 | 0.037 | 0.4072 | No |
| 208 | PLCG2 | NA | 27591 | 0.035 | 0.4019 | No |
| 209 | MIR548P | NA | 28068 | 0.032 | 0.3936 | No |
| 210 | DPM1 | NA | 28550 | 0.029 | 0.3852 | No |
| 211 | GPAM | NA | 28801 | 0.027 | 0.3810 | No |
| 212 | TAZ | NA | 28909 | 0.027 | 0.3795 | No |
| 213 | MTMR2 | NA | 29851 | 0.021 | 0.3625 | No |
| 214 | INPP5E | NA | 30241 | 0.019 | 0.3556 | No |
| 215 | PLEKHA2 | NA | 31306 | 0.014 | 0.3363 | No |
| 216 | PLA2G2A | NA | 31517 | 0.013 | 0.3326 | No |
| 217 | CPNE1 | NA | 32664 | 0.008 | 0.3117 | No |
| 218 | PIGH | NA | 33069 | 0.007 | 0.3043 | No |
| 219 | NR1H2 | NA | 33341 | 0.005 | 0.2994 | No |
| 220 | PLA2G2F | NA | 33945 | 0.001 | 0.2883 | No |
| 221 | MIR30C1 | NA | 36230 | 0.000 | 0.2463 | No |
| 222 | TAMM41 | NA | 39464 | -0.005 | 0.1869 | No |
| 223 | PLA2G2D | NA | 40056 | -0.012 | 0.1762 | No |
| 224 | HTR2A | NA | 40373 | -0.016 | 0.1706 | No |
| 225 | DGAT2L6 | NA | 40401 | -0.017 | 0.1704 | No |
| 226 | TPTE2 | NA | 41312 | -0.027 | 0.1541 | No |
| 227 | CHAT | NA | 41704 | -0.032 | 0.1474 | No |
| 228 | PLA2G5 | NA | 42850 | -0.047 | 0.1270 | No |
| 229 | DGAT2 | NA | 42859 | -0.047 | 0.1276 | No |
| 230 | C3 | NA | 43139 | -0.050 | 0.1233 | No |
| 231 | ACHE | NA | 43273 | -0.052 | 0.1217 | No |
| 232 | PLA2G4C | NA | 43475 | -0.055 | 0.1188 | No |
| 233 | FABP5 | NA | 43523 | -0.055 | 0.1188 | No |
| 234 | ACSL4 | NA | 43957 | -0.061 | 0.1118 | No |
| 235 | LPCAT1 | NA | 44007 | -0.062 | 0.1119 | No |
| 236 | PNPLA3 | NA | 45113 | -0.078 | 0.0928 | No |
| 237 | LPCAT4 | NA | 45175 | -0.079 | 0.0929 | No |
| 238 | PITPNM2 | NA | 45598 | -0.085 | 0.0865 | No |
| 239 | ARF1 | NA | 46706 | -0.102 | 0.0677 | No |
| 240 | NR1H3 | NA | 47515 | -0.117 | 0.0547 | No |
| 241 | LDLR | NA | 47518 | -0.117 | 0.0565 | No |
| 242 | HTR2C | NA | 47543 | -0.117 | 0.0579 | No |
| 243 | AGMO | NA | 47701 | -0.120 | 0.0569 | No |
| 244 | ETNPPL | NA | 47794 | -0.122 | 0.0571 | No |
| 245 | PHOSPHO1 | NA | 48535 | -0.135 | 0.0456 | No |
| 246 | PLEKHA4 | NA | 48933 | -0.143 | 0.0406 | No |
| 247 | DPM3 | NA | 48935 | -0.143 | 0.0428 | No |
| 248 | LPCAT3 | NA | 49243 | -0.150 | 0.0395 | No |
| 249 | THRSP | NA | 49690 | -0.159 | 0.0338 | No |
| 250 | PDGFB | NA | 49822 | -0.162 | 0.0340 | No |
| 251 | CSNK2B | NA | 49931 | -0.165 | 0.0346 | No |
| 252 | PGS1 | NA | 50423 | -0.175 | 0.0283 | No |
| 253 | APOA2 | NA | 50997 | -0.189 | 0.0208 | No |
| 254 | MTMR8 | NA | 51205 | -0.194 | 0.0200 | No |
| 255 | PIGA | NA | 51410 | -0.200 | 0.0194 | No |
| 256 | PYURF | NA | 51900 | -0.212 | 0.0138 | No |
| 257 | APOA1 | NA | 51971 | -0.214 | 0.0159 | No |
| 258 | BMX | NA | 53116 | -0.251 | -0.0012 | No |
| 259 | SCARB1 | NA | 53302 | -0.261 | -0.0005 | No |
| 260 | DGKB | NA | 53337 | -0.263 | 0.0030 | No |
| 261 | INPP5D | NA | 54339 | -0.407 | -0.0090 | No |
| 262 | PLSCR1 | NA | 54364 | -0.417 | -0.0028 | No |
| 263 | CPNE6 | NA | 54424 | -0.444 | 0.0031 | No |