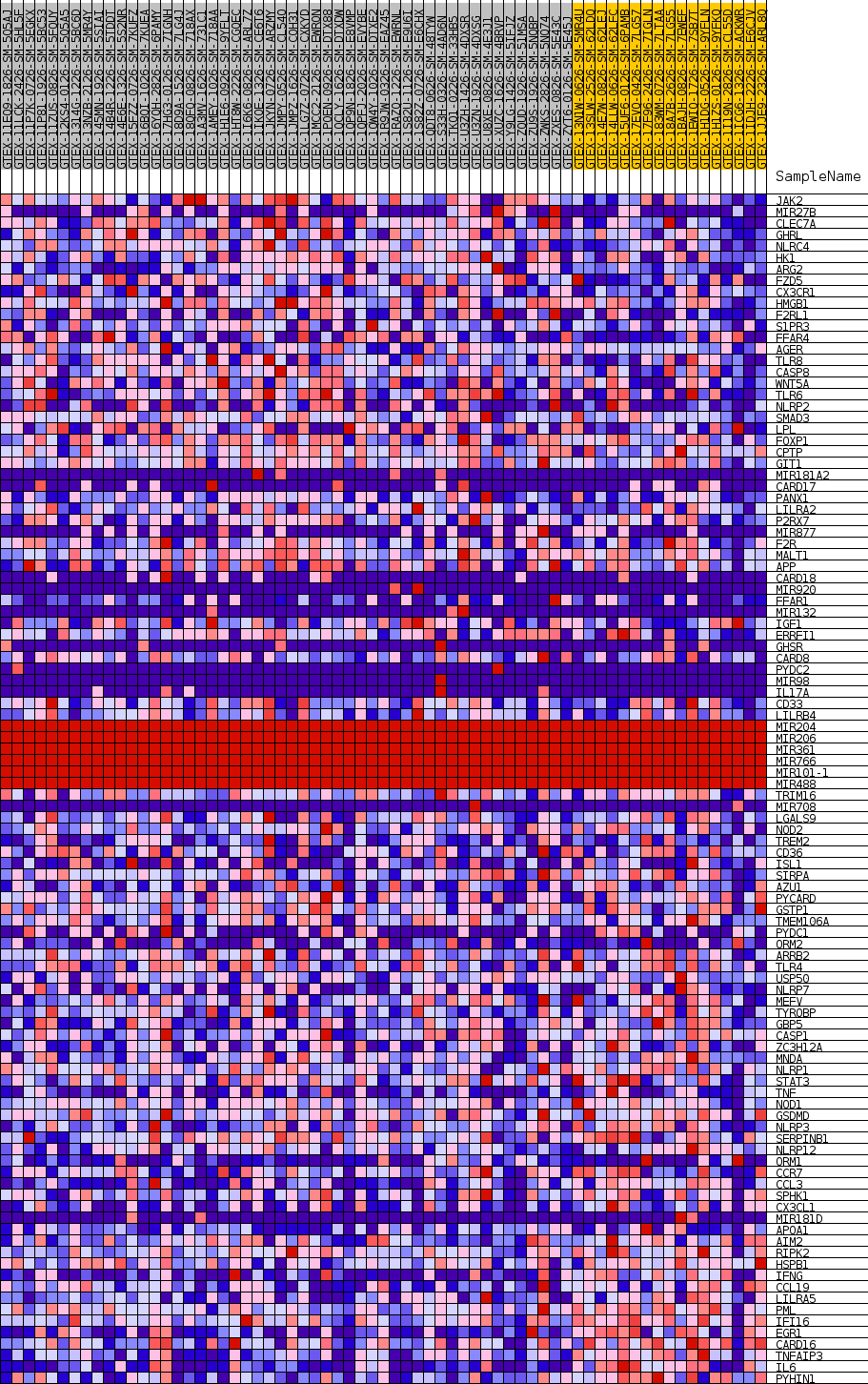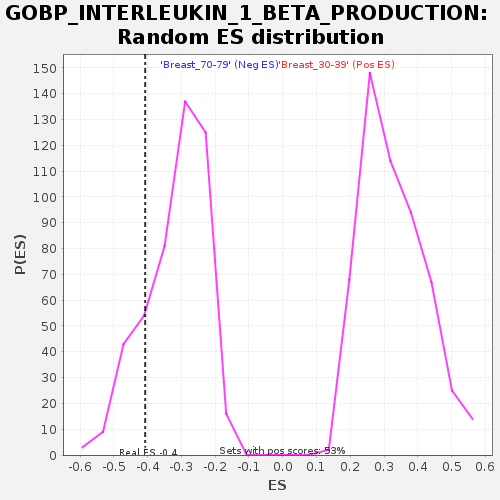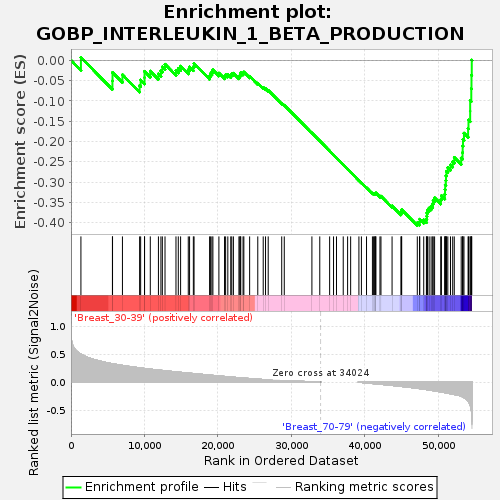
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Breast.Breast.cls #Breast_30-39_versus_Breast_70-79.Breast.cls #Breast_30-39_versus_Breast_70-79_repos |
| Phenotype | Breast.cls#Breast_30-39_versus_Breast_70-79_repos |
| Upregulated in class | Breast_70-79 |
| GeneSet | GOBP_INTERLEUKIN_1_BETA_PRODUCTION |
| Enrichment Score (ES) | -0.40705368 |
| Normalized Enrichment Score (NES) | -1.280588 |
| Nominal p-value | 0.18162394 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.998 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | JAK2 | NA | 1350 | 0.499 | 0.0071 | No |
| 2 | MIR27B | NA | 5640 | 0.329 | -0.0506 | No |
| 3 | CLEC7A | NA | 5655 | 0.329 | -0.0299 | No |
| 4 | GHRL | NA | 7003 | 0.299 | -0.0356 | No |
| 5 | NLRC4 | NA | 9342 | 0.255 | -0.0622 | No |
| 6 | HK1 | NA | 9483 | 0.253 | -0.0486 | No |
| 7 | ARG2 | NA | 10010 | 0.244 | -0.0427 | No |
| 8 | FZD5 | NA | 10015 | 0.244 | -0.0271 | No |
| 9 | CX3CR1 | NA | 10796 | 0.232 | -0.0267 | No |
| 10 | HMGB1 | NA | 11903 | 0.216 | -0.0332 | No |
| 11 | F2RL1 | NA | 12244 | 0.211 | -0.0259 | No |
| 12 | S1PR3 | NA | 12436 | 0.209 | -0.0161 | No |
| 13 | FFAR4 | NA | 12800 | 0.204 | -0.0098 | No |
| 14 | AGER | NA | 14292 | 0.183 | -0.0255 | No |
| 15 | TLR8 | NA | 14608 | 0.179 | -0.0198 | No |
| 16 | CASP8 | NA | 14923 | 0.175 | -0.0144 | No |
| 17 | WNT5A | NA | 15967 | 0.162 | -0.0233 | No |
| 18 | TLR6 | NA | 16144 | 0.160 | -0.0163 | No |
| 19 | NLRP2 | NA | 16657 | 0.153 | -0.0159 | No |
| 20 | SMAD3 | NA | 16754 | 0.152 | -0.0079 | No |
| 21 | LPL | NA | 18871 | 0.125 | -0.0388 | No |
| 22 | FOXP1 | NA | 18979 | 0.124 | -0.0328 | No |
| 23 | CPTP | NA | 19179 | 0.121 | -0.0287 | No |
| 24 | GIT1 | NA | 19319 | 0.120 | -0.0236 | No |
| 25 | MIR181A2 | NA | 20136 | 0.110 | -0.0316 | No |
| 26 | CARD17 | NA | 20904 | 0.101 | -0.0392 | No |
| 27 | PANX1 | NA | 21051 | 0.099 | -0.0356 | No |
| 28 | LILRA2 | NA | 21352 | 0.095 | -0.0350 | No |
| 29 | P2RX7 | NA | 21752 | 0.091 | -0.0366 | No |
| 30 | MIR877 | NA | 21890 | 0.089 | -0.0334 | No |
| 31 | F2R | NA | 22111 | 0.087 | -0.0318 | No |
| 32 | MALT1 | NA | 22866 | 0.079 | -0.0407 | No |
| 33 | APP | NA | 22986 | 0.077 | -0.0379 | No |
| 34 | CARD18 | NA | 23047 | 0.077 | -0.0341 | No |
| 35 | MIR920 | NA | 23114 | 0.076 | -0.0305 | No |
| 36 | FFAR1 | NA | 23425 | 0.073 | -0.0315 | No |
| 37 | MIR132 | NA | 23534 | 0.072 | -0.0289 | No |
| 38 | IGF1 | NA | 24322 | 0.064 | -0.0393 | No |
| 39 | ERRFI1 | NA | 25430 | 0.053 | -0.0562 | No |
| 40 | GHSR | NA | 26170 | 0.046 | -0.0668 | No |
| 41 | CARD8 | NA | 26486 | 0.043 | -0.0699 | No |
| 42 | PYDC2 | NA | 26850 | 0.040 | -0.0739 | No |
| 43 | MIR98 | NA | 28690 | 0.028 | -0.1059 | No |
| 44 | IL17A | NA | 29037 | 0.026 | -0.1106 | No |
| 45 | CD33 | NA | 32800 | 0.008 | -0.1791 | No |
| 46 | LILRB4 | NA | 33871 | 0.002 | -0.1986 | No |
| 47 | MIR204 | NA | 35214 | 0.000 | -0.2233 | No |
| 48 | MIR206 | NA | 35744 | 0.000 | -0.2330 | No |
| 49 | MIR361 | NA | 36147 | 0.000 | -0.2404 | No |
| 50 | MIR766 | NA | 37065 | 0.000 | -0.2572 | No |
| 51 | MIR101-1 | NA | 37661 | 0.000 | -0.2681 | No |
| 52 | MIR488 | NA | 38075 | 0.000 | -0.2757 | No |
| 53 | TRIM16 | NA | 39199 | -0.001 | -0.2962 | No |
| 54 | MIR708 | NA | 39539 | -0.005 | -0.3021 | No |
| 55 | LGALS9 | NA | 40236 | -0.015 | -0.3139 | No |
| 56 | NOD2 | NA | 41039 | -0.024 | -0.3271 | No |
| 57 | TREM2 | NA | 41202 | -0.026 | -0.3284 | No |
| 58 | CD36 | NA | 41299 | -0.027 | -0.3284 | No |
| 59 | ISL1 | NA | 41392 | -0.028 | -0.3283 | No |
| 60 | SIRPA | NA | 41470 | -0.029 | -0.3279 | No |
| 61 | AZU1 | NA | 41510 | -0.030 | -0.3267 | No |
| 62 | PYCARD | NA | 42066 | -0.036 | -0.3346 | No |
| 63 | GSTP1 | NA | 42202 | -0.038 | -0.3346 | No |
| 64 | TMEM106A | NA | 43714 | -0.058 | -0.3587 | No |
| 65 | PYDC1 | NA | 44921 | -0.075 | -0.3760 | No |
| 66 | ORM2 | NA | 44995 | -0.076 | -0.3725 | No |
| 67 | ARRB2 | NA | 45025 | -0.077 | -0.3681 | No |
| 68 | TLR4 | NA | 47149 | -0.110 | -0.4000 | Yes |
| 69 | USP50 | NA | 47458 | -0.115 | -0.3983 | Yes |
| 70 | NLRP7 | NA | 47482 | -0.116 | -0.3913 | Yes |
| 71 | MEFV | NA | 48024 | -0.126 | -0.3932 | Yes |
| 72 | TYROBP | NA | 48389 | -0.132 | -0.3914 | Yes |
| 73 | GBP5 | NA | 48445 | -0.134 | -0.3839 | Yes |
| 74 | CASP1 | NA | 48465 | -0.134 | -0.3757 | Yes |
| 75 | ZC3H12A | NA | 48550 | -0.136 | -0.3685 | Yes |
| 76 | MNDA | NA | 48763 | -0.140 | -0.3635 | Yes |
| 77 | NLRP1 | NA | 49029 | -0.145 | -0.3591 | Yes |
| 78 | STAT3 | NA | 49249 | -0.150 | -0.3536 | Yes |
| 79 | TNF | NA | 49309 | -0.151 | -0.3450 | Yes |
| 80 | NOD1 | NA | 49505 | -0.155 | -0.3387 | Yes |
| 81 | GSDMD | NA | 50341 | -0.174 | -0.3429 | Yes |
| 82 | NLRP3 | NA | 50431 | -0.176 | -0.3333 | Yes |
| 83 | SERPINB1 | NA | 50895 | -0.187 | -0.3299 | Yes |
| 84 | NLRP12 | NA | 50927 | -0.188 | -0.3185 | Yes |
| 85 | ORM1 | NA | 50951 | -0.188 | -0.3069 | Yes |
| 86 | CCR7 | NA | 51037 | -0.190 | -0.2963 | Yes |
| 87 | CCL3 | NA | 51058 | -0.191 | -0.2845 | Yes |
| 88 | SPHK1 | NA | 51119 | -0.192 | -0.2733 | Yes |
| 89 | CX3CL1 | NA | 51304 | -0.197 | -0.2641 | Yes |
| 90 | MIR181D | NA | 51672 | -0.207 | -0.2577 | Yes |
| 91 | APOA1 | NA | 51971 | -0.214 | -0.2495 | Yes |
| 92 | AIM2 | NA | 52182 | -0.220 | -0.2393 | Yes |
| 93 | RIPK2 | NA | 53126 | -0.251 | -0.2405 | Yes |
| 94 | HSPB1 | NA | 53310 | -0.262 | -0.2272 | Yes |
| 95 | IFNG | NA | 53327 | -0.263 | -0.2107 | Yes |
| 96 | CCL19 | NA | 53370 | -0.266 | -0.1945 | Yes |
| 97 | LILRA5 | NA | 53504 | -0.275 | -0.1794 | Yes |
| 98 | PML | NA | 54066 | -0.335 | -0.1683 | Yes |
| 99 | IFI16 | NA | 54119 | -0.346 | -0.1471 | Yes |
| 100 | EGR1 | NA | 54347 | -0.409 | -0.1252 | Yes |
| 101 | CARD16 | NA | 54349 | -0.410 | -0.0990 | Yes |
| 102 | TNFAIP3 | NA | 54500 | -0.501 | -0.0698 | Yes |
| 103 | IL6 | NA | 54527 | -0.525 | -0.0367 | Yes |
| 104 | PYHIN1 | NA | 54564 | -0.593 | 0.0005 | Yes |