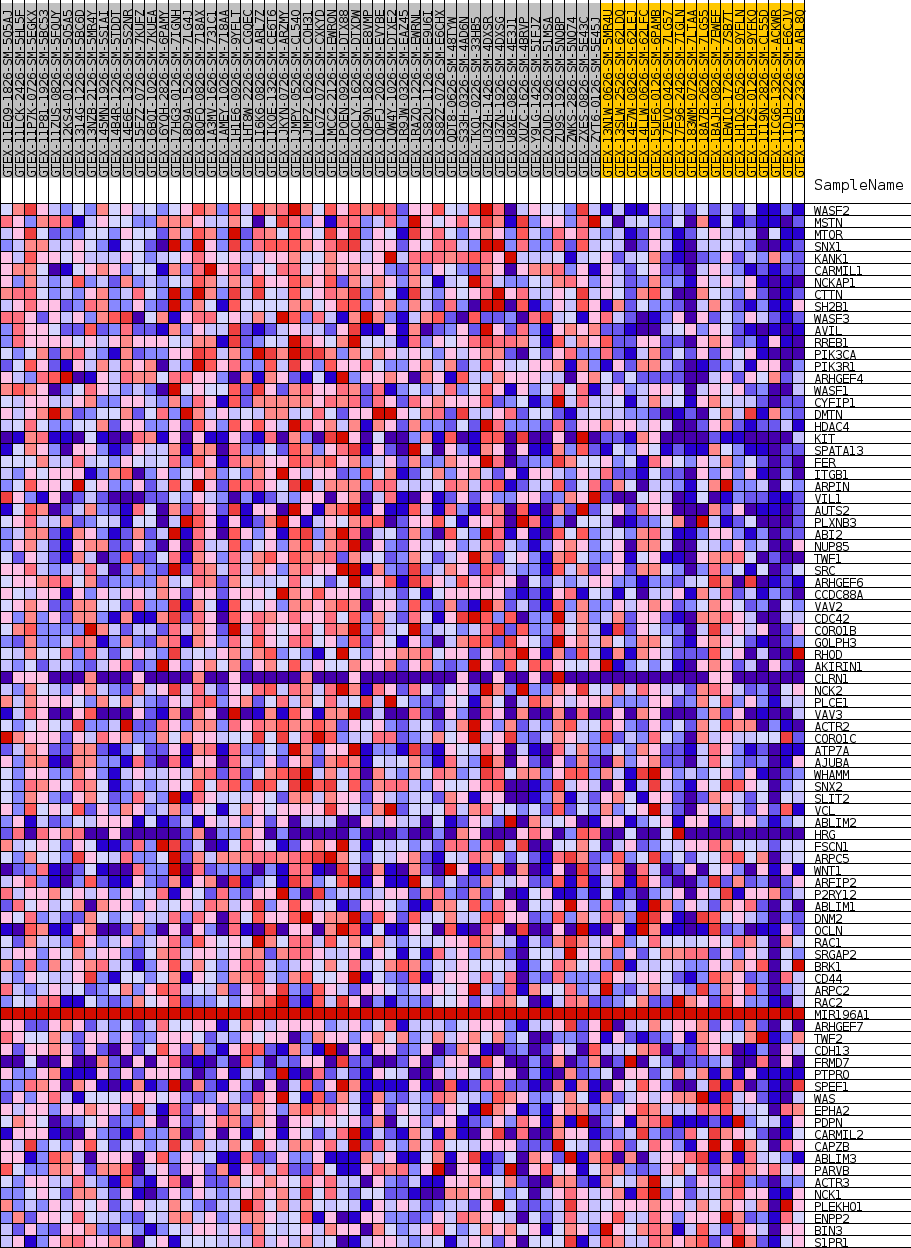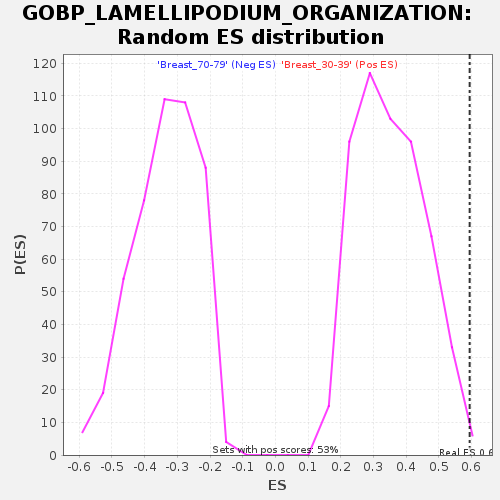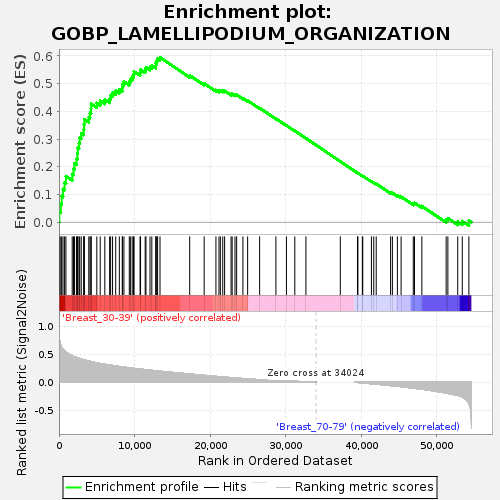
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Breast.Breast.cls #Breast_30-39_versus_Breast_70-79.Breast.cls #Breast_30-39_versus_Breast_70-79_repos |
| Phenotype | Breast.cls#Breast_30-39_versus_Breast_70-79_repos |
| Upregulated in class | Breast_30-39 |
| GeneSet | GOBP_LAMELLIPODIUM_ORGANIZATION |
| Enrichment Score (ES) | 0.59443206 |
| Normalized Enrichment Score (NES) | 1.6892183 |
| Nominal p-value | 0.005628518 |
| FDR q-value | 0.302837 |
| FWER p-Value | 0.686 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | WASF2 | NA | 37 | 0.784 | 0.0375 | Yes |
| 2 | MSTN | NA | 238 | 0.657 | 0.0658 | Yes |
| 3 | MTOR | NA | 355 | 0.623 | 0.0940 | Yes |
| 4 | SNX1 | NA | 530 | 0.588 | 0.1194 | Yes |
| 5 | KANK1 | NA | 740 | 0.558 | 0.1427 | Yes |
| 6 | CARMIL1 | NA | 916 | 0.539 | 0.1657 | Yes |
| 7 | NCKAP1 | NA | 1748 | 0.473 | 0.1735 | Yes |
| 8 | CTTN | NA | 1923 | 0.460 | 0.1927 | Yes |
| 9 | SH2B1 | NA | 2060 | 0.453 | 0.2122 | Yes |
| 10 | WASF3 | NA | 2321 | 0.439 | 0.2288 | Yes |
| 11 | AVIL | NA | 2444 | 0.433 | 0.2477 | Yes |
| 12 | RREB1 | NA | 2478 | 0.432 | 0.2681 | Yes |
| 13 | PIK3CA | NA | 2652 | 0.424 | 0.2856 | Yes |
| 14 | PIK3R1 | NA | 2739 | 0.420 | 0.3044 | Yes |
| 15 | ARHGEF4 | NA | 2943 | 0.411 | 0.3207 | Yes |
| 16 | WASF1 | NA | 3273 | 0.399 | 0.3341 | Yes |
| 17 | CYFIP1 | NA | 3310 | 0.398 | 0.3528 | Yes |
| 18 | DMTN | NA | 3358 | 0.396 | 0.3712 | Yes |
| 19 | HDAC4 | NA | 3944 | 0.376 | 0.3788 | Yes |
| 20 | KIT | NA | 4102 | 0.371 | 0.3939 | Yes |
| 21 | SPATA13 | NA | 4241 | 0.367 | 0.4093 | Yes |
| 22 | FER | NA | 4254 | 0.367 | 0.4269 | Yes |
| 23 | ITGB1 | NA | 5005 | 0.345 | 0.4299 | Yes |
| 24 | ARPIN | NA | 5456 | 0.333 | 0.4379 | Yes |
| 25 | VIL1 | NA | 6064 | 0.319 | 0.4423 | Yes |
| 26 | AUTS2 | NA | 6705 | 0.305 | 0.4453 | Yes |
| 27 | PLXNB3 | NA | 6832 | 0.302 | 0.4578 | Yes |
| 28 | ABI2 | NA | 7092 | 0.297 | 0.4675 | Yes |
| 29 | NUP85 | NA | 7507 | 0.289 | 0.4739 | Yes |
| 30 | TWF1 | NA | 7986 | 0.279 | 0.4787 | Yes |
| 31 | SRC | NA | 8379 | 0.272 | 0.4848 | Yes |
| 32 | ARHGEF6 | NA | 8398 | 0.272 | 0.4977 | Yes |
| 33 | CCDC88A | NA | 8595 | 0.268 | 0.5072 | Yes |
| 34 | VAV2 | NA | 9305 | 0.256 | 0.5066 | Yes |
| 35 | CDC42 | NA | 9447 | 0.254 | 0.5163 | Yes |
| 36 | CORO1B | NA | 9687 | 0.249 | 0.5241 | Yes |
| 37 | GOLPH3 | NA | 9864 | 0.247 | 0.5329 | Yes |
| 38 | RHOD | NA | 9937 | 0.245 | 0.5435 | Yes |
| 39 | AKIRIN1 | NA | 10732 | 0.233 | 0.5403 | Yes |
| 40 | CLRN1 | NA | 10797 | 0.232 | 0.5504 | Yes |
| 41 | NCK2 | NA | 11409 | 0.223 | 0.5500 | Yes |
| 42 | PLCE1 | NA | 11513 | 0.221 | 0.5589 | Yes |
| 43 | VAV3 | NA | 12048 | 0.214 | 0.5595 | Yes |
| 44 | ACTR2 | NA | 12288 | 0.211 | 0.5654 | Yes |
| 45 | CORO1C | NA | 12817 | 0.203 | 0.5656 | Yes |
| 46 | ATP7A | NA | 12830 | 0.203 | 0.5753 | Yes |
| 47 | AJUBA | NA | 12946 | 0.201 | 0.5830 | Yes |
| 48 | WHAMM | NA | 13054 | 0.200 | 0.5907 | Yes |
| 49 | SNX2 | NA | 13371 | 0.195 | 0.5944 | Yes |
| 50 | SLIT2 | NA | 17302 | 0.145 | 0.5294 | No |
| 51 | VCL | NA | 19217 | 0.121 | 0.5002 | No |
| 52 | ABLIM2 | NA | 20775 | 0.102 | 0.4766 | No |
| 53 | HRG | NA | 21179 | 0.098 | 0.4739 | No |
| 54 | FSCN1 | NA | 21359 | 0.095 | 0.4753 | No |
| 55 | ARPC5 | NA | 21676 | 0.092 | 0.4740 | No |
| 56 | WNT1 | NA | 21924 | 0.089 | 0.4738 | No |
| 57 | ARFIP2 | NA | 22778 | 0.080 | 0.4620 | No |
| 58 | P2RY12 | NA | 22922 | 0.078 | 0.4632 | No |
| 59 | ABLIM1 | NA | 23296 | 0.074 | 0.4599 | No |
| 60 | DNM2 | NA | 23515 | 0.072 | 0.4594 | No |
| 61 | OCLN | NA | 24346 | 0.064 | 0.4473 | No |
| 62 | RAC1 | NA | 24971 | 0.058 | 0.4386 | No |
| 63 | SRGAP2 | NA | 26561 | 0.043 | 0.4116 | No |
| 64 | BRK1 | NA | 28713 | 0.028 | 0.3735 | No |
| 65 | CD44 | NA | 30107 | 0.020 | 0.3489 | No |
| 66 | ARPC2 | NA | 31215 | 0.014 | 0.3292 | No |
| 67 | RAC2 | NA | 32687 | 0.008 | 0.3027 | No |
| 68 | MIR196A1 | NA | 37238 | 0.000 | 0.2192 | No |
| 69 | ARHGEF7 | NA | 39512 | -0.005 | 0.1777 | No |
| 70 | TWF2 | NA | 39570 | -0.006 | 0.1770 | No |
| 71 | CDH13 | NA | 40155 | -0.014 | 0.1669 | No |
| 72 | FRMD7 | NA | 40211 | -0.014 | 0.1666 | No |
| 73 | PTPRO | NA | 41358 | -0.028 | 0.1469 | No |
| 74 | SPEF1 | NA | 41667 | -0.031 | 0.1428 | No |
| 75 | WAS | NA | 41993 | -0.035 | 0.1385 | No |
| 76 | EPHA2 | NA | 43897 | -0.061 | 0.1066 | No |
| 77 | PDPN | NA | 44133 | -0.064 | 0.1054 | No |
| 78 | CARMIL2 | NA | 44795 | -0.073 | 0.0968 | No |
| 79 | CAPZB | NA | 45287 | -0.080 | 0.0917 | No |
| 80 | ABLIM3 | NA | 46900 | -0.106 | 0.0673 | No |
| 81 | PARVB | NA | 47059 | -0.108 | 0.0697 | No |
| 82 | ACTR3 | NA | 48033 | -0.126 | 0.0579 | No |
| 83 | NCK1 | NA | 51252 | -0.196 | 0.0084 | No |
| 84 | PLEKHO1 | NA | 51479 | -0.201 | 0.0141 | No |
| 85 | ENPP2 | NA | 52783 | -0.237 | 0.0017 | No |
| 86 | BIN3 | NA | 53384 | -0.266 | 0.0037 | No |
| 87 | S1PR1 | NA | 54260 | -0.379 | 0.0061 | No |