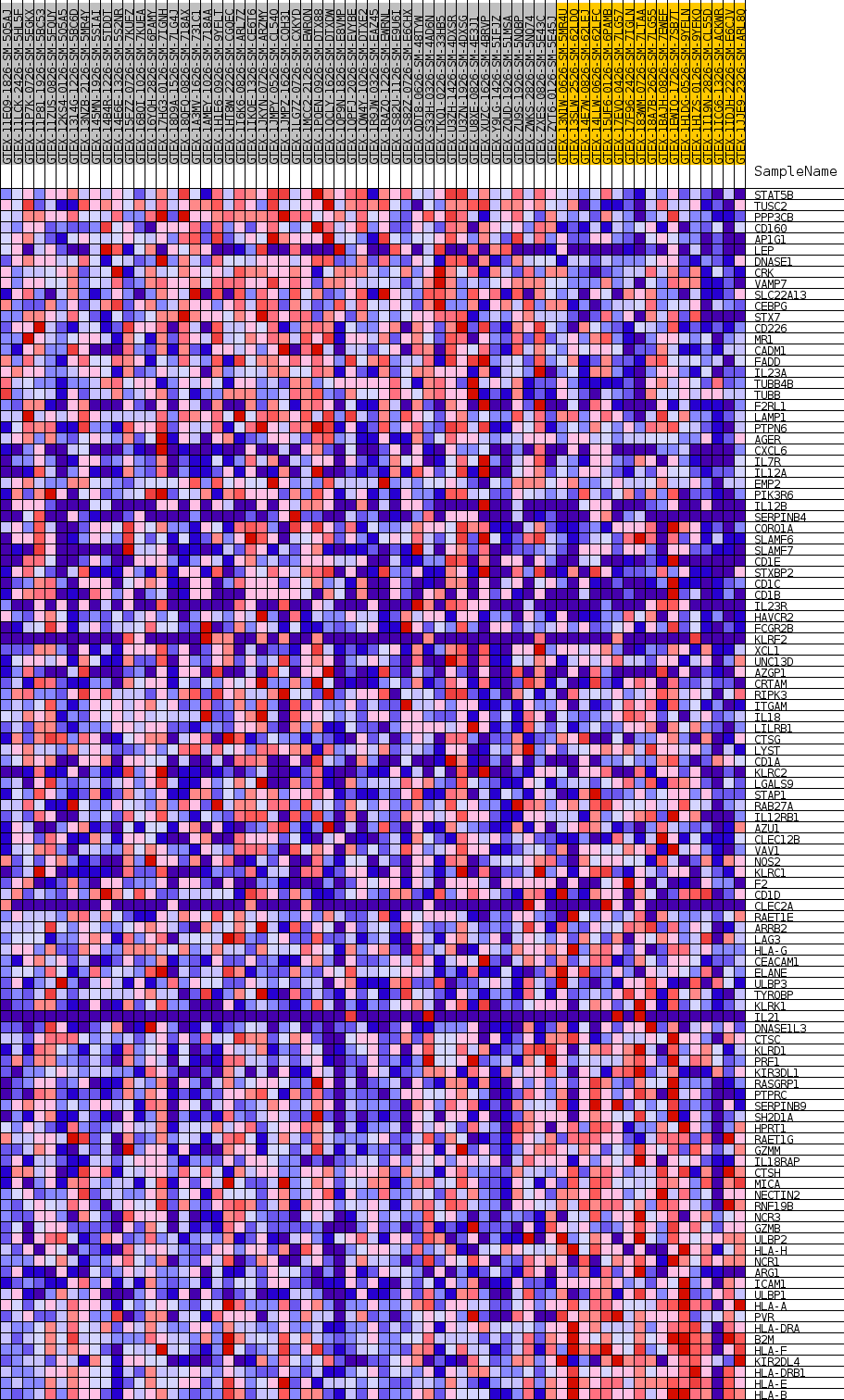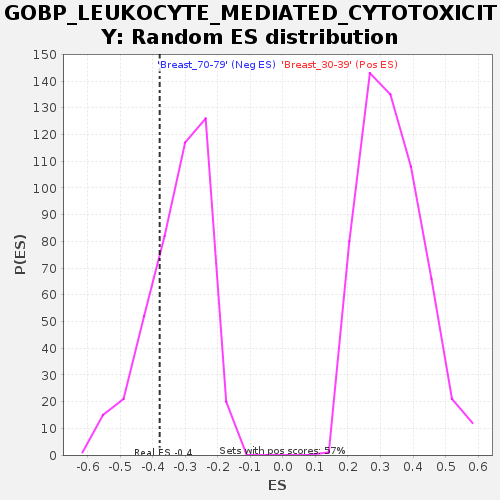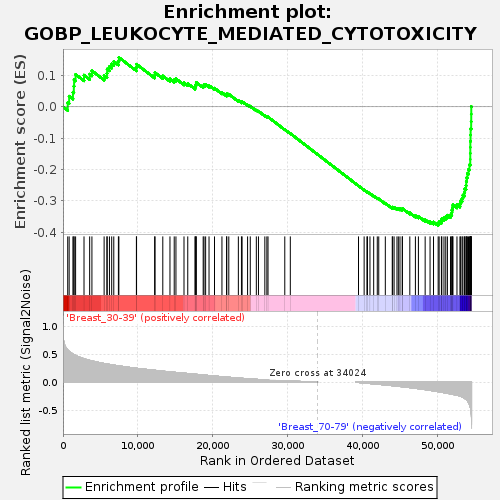
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Breast.Breast.cls #Breast_30-39_versus_Breast_70-79.Breast.cls #Breast_30-39_versus_Breast_70-79_repos |
| Phenotype | Breast.cls#Breast_30-39_versus_Breast_70-79_repos |
| Upregulated in class | Breast_70-79 |
| GeneSet | GOBP_LEUKOCYTE_MEDIATED_CYTOTOXICITY |
| Enrichment Score (ES) | -0.37921372 |
| Normalized Enrichment Score (NES) | -1.1766562 |
| Nominal p-value | 0.25345623 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.999 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | STAT5B | NA | 632 | 0.572 | 0.0133 | No |
| 2 | TUSC2 | NA | 822 | 0.548 | 0.0338 | No |
| 3 | PPP3CB | NA | 1327 | 0.501 | 0.0464 | No |
| 4 | CD160 | NA | 1447 | 0.492 | 0.0656 | No |
| 5 | AP1G1 | NA | 1483 | 0.489 | 0.0863 | No |
| 6 | LEP | NA | 1684 | 0.477 | 0.1034 | No |
| 7 | DNASE1 | NA | 2807 | 0.417 | 0.1010 | No |
| 8 | CRK | NA | 3562 | 0.389 | 0.1042 | No |
| 9 | VAMP7 | NA | 3864 | 0.378 | 0.1151 | No |
| 10 | SLC22A13 | NA | 5494 | 0.332 | 0.0997 | No |
| 11 | CEBPG | NA | 5871 | 0.323 | 0.1069 | No |
| 12 | STX7 | NA | 5899 | 0.323 | 0.1205 | No |
| 13 | CD226 | NA | 6206 | 0.316 | 0.1287 | No |
| 14 | MR1 | NA | 6506 | 0.309 | 0.1366 | No |
| 15 | CADM1 | NA | 6785 | 0.303 | 0.1448 | No |
| 16 | FADD | NA | 7402 | 0.291 | 0.1461 | No |
| 17 | IL23A | NA | 7465 | 0.290 | 0.1576 | No |
| 18 | TUBB4B | NA | 9807 | 0.248 | 0.1254 | No |
| 19 | TUBB | NA | 9829 | 0.247 | 0.1358 | No |
| 20 | F2RL1 | NA | 12244 | 0.211 | 0.1008 | No |
| 21 | LAMP1 | NA | 12298 | 0.211 | 0.1090 | No |
| 22 | PTPN6 | NA | 13334 | 0.196 | 0.0985 | No |
| 23 | AGER | NA | 14292 | 0.183 | 0.0889 | No |
| 24 | CXCL6 | NA | 14860 | 0.176 | 0.0862 | No |
| 25 | IL7R | NA | 15091 | 0.173 | 0.0895 | No |
| 26 | IL12A | NA | 16167 | 0.159 | 0.0767 | No |
| 27 | EMP2 | NA | 16669 | 0.153 | 0.0742 | No |
| 28 | PIK3R6 | NA | 17640 | 0.141 | 0.0625 | No |
| 29 | IL12B | NA | 17687 | 0.141 | 0.0678 | No |
| 30 | SERPINB4 | NA | 17772 | 0.140 | 0.0724 | No |
| 31 | CORO1A | NA | 17805 | 0.139 | 0.0779 | No |
| 32 | SLAMF6 | NA | 18730 | 0.127 | 0.0664 | No |
| 33 | SLAMF7 | NA | 18841 | 0.126 | 0.0699 | No |
| 34 | CD1E | NA | 19058 | 0.123 | 0.0713 | No |
| 35 | STXBP2 | NA | 19511 | 0.117 | 0.0681 | No |
| 36 | CD1C | NA | 20243 | 0.108 | 0.0594 | No |
| 37 | CD1B | NA | 21237 | 0.097 | 0.0454 | No |
| 38 | IL23R | NA | 21868 | 0.090 | 0.0378 | No |
| 39 | HAVCR2 | NA | 21869 | 0.090 | 0.0417 | No |
| 40 | FCGR2B | NA | 22153 | 0.087 | 0.0402 | No |
| 41 | KLRF2 | NA | 23430 | 0.073 | 0.0200 | No |
| 42 | XCL1 | NA | 23843 | 0.068 | 0.0154 | No |
| 43 | UNC13D | NA | 23928 | 0.068 | 0.0168 | No |
| 44 | AZGP1 | NA | 24663 | 0.060 | 0.0060 | No |
| 45 | CRTAM | NA | 25021 | 0.057 | 0.0019 | No |
| 46 | RIPK3 | NA | 25831 | 0.049 | -0.0108 | No |
| 47 | ITGAM | NA | 26116 | 0.047 | -0.0139 | No |
| 48 | IL18 | NA | 26968 | 0.040 | -0.0278 | No |
| 49 | LILRB1 | NA | 27240 | 0.038 | -0.0312 | No |
| 50 | CTSG | NA | 27392 | 0.037 | -0.0323 | No |
| 51 | LYST | NA | 29633 | 0.022 | -0.0725 | No |
| 52 | CD1A | NA | 30372 | 0.018 | -0.0852 | No |
| 53 | KLRC2 | NA | 39484 | -0.005 | -0.2522 | No |
| 54 | LGALS9 | NA | 40236 | -0.015 | -0.2654 | No |
| 55 | STAP1 | NA | 40610 | -0.019 | -0.2714 | No |
| 56 | RAB27A | NA | 40682 | -0.020 | -0.2718 | No |
| 57 | IL12RB1 | NA | 41014 | -0.024 | -0.2769 | No |
| 58 | AZU1 | NA | 41510 | -0.030 | -0.2847 | No |
| 59 | CLEC12B | NA | 41992 | -0.035 | -0.2920 | No |
| 60 | VAV1 | NA | 42188 | -0.038 | -0.2939 | No |
| 61 | NOS2 | NA | 43066 | -0.049 | -0.3078 | No |
| 62 | KLRC1 | NA | 43992 | -0.062 | -0.3221 | No |
| 63 | F2 | NA | 44044 | -0.062 | -0.3203 | No |
| 64 | CD1D | NA | 44239 | -0.065 | -0.3210 | No |
| 65 | CLEC2A | NA | 44623 | -0.071 | -0.3250 | No |
| 66 | RAET1E | NA | 44805 | -0.073 | -0.3251 | No |
| 67 | ARRB2 | NA | 45025 | -0.077 | -0.3258 | No |
| 68 | LAG3 | NA | 45326 | -0.081 | -0.3277 | No |
| 69 | HLA-G | NA | 45353 | -0.081 | -0.3247 | No |
| 70 | CEACAM1 | NA | 46340 | -0.097 | -0.3385 | No |
| 71 | ELANE | NA | 47085 | -0.109 | -0.3475 | No |
| 72 | ULBP3 | NA | 47487 | -0.116 | -0.3498 | No |
| 73 | TYROBP | NA | 48389 | -0.132 | -0.3605 | No |
| 74 | KLRK1 | NA | 49043 | -0.146 | -0.3661 | No |
| 75 | IL21 | NA | 49506 | -0.155 | -0.3679 | No |
| 76 | DNASE1L3 | NA | 50126 | -0.169 | -0.3718 | Yes |
| 77 | CTSC | NA | 50256 | -0.172 | -0.3667 | Yes |
| 78 | KLRD1 | NA | 50561 | -0.178 | -0.3645 | Yes |
| 79 | PRF1 | NA | 50616 | -0.179 | -0.3577 | Yes |
| 80 | KIR3DL1 | NA | 50903 | -0.187 | -0.3548 | Yes |
| 81 | RASGRP1 | NA | 51143 | -0.193 | -0.3508 | Yes |
| 82 | PTPRC | NA | 51361 | -0.198 | -0.3461 | Yes |
| 83 | SERPINB9 | NA | 51773 | -0.209 | -0.3445 | Yes |
| 84 | SH2D1A | NA | 51946 | -0.213 | -0.3384 | Yes |
| 85 | HPRT1 | NA | 51964 | -0.214 | -0.3294 | Yes |
| 86 | RAET1G | NA | 52044 | -0.216 | -0.3214 | Yes |
| 87 | GZMM | NA | 52105 | -0.218 | -0.3130 | Yes |
| 88 | IL18RAP | NA | 52639 | -0.233 | -0.3126 | Yes |
| 89 | CTSH | NA | 53038 | -0.247 | -0.3091 | Yes |
| 90 | MICA | NA | 53128 | -0.251 | -0.2998 | Yes |
| 91 | NECTIN2 | NA | 53322 | -0.262 | -0.2919 | Yes |
| 92 | RNF19B | NA | 53427 | -0.270 | -0.2821 | Yes |
| 93 | NCR3 | NA | 53635 | -0.286 | -0.2734 | Yes |
| 94 | GZMB | NA | 53679 | -0.290 | -0.2615 | Yes |
| 95 | ULBP2 | NA | 53851 | -0.306 | -0.2513 | Yes |
| 96 | HLA-H | NA | 53882 | -0.309 | -0.2384 | Yes |
| 97 | NCR1 | NA | 53943 | -0.317 | -0.2257 | Yes |
| 98 | ARG1 | NA | 54035 | -0.331 | -0.2129 | Yes |
| 99 | ICAM1 | NA | 54181 | -0.359 | -0.2000 | Yes |
| 100 | ULBP1 | NA | 54305 | -0.394 | -0.1850 | Yes |
| 101 | HLA-A | NA | 54414 | -0.439 | -0.1679 | Yes |
| 102 | PVR | NA | 54417 | -0.441 | -0.1487 | Yes |
| 103 | HLA-DRA | NA | 54422 | -0.444 | -0.1294 | Yes |
| 104 | B2M | NA | 54426 | -0.445 | -0.1100 | Yes |
| 105 | HLA-F | NA | 54455 | -0.467 | -0.0902 | Yes |
| 106 | KIR2DL4 | NA | 54465 | -0.472 | -0.0697 | Yes |
| 107 | HLA-DRB1 | NA | 54537 | -0.541 | -0.0474 | Yes |
| 108 | HLA-E | NA | 54547 | -0.553 | -0.0235 | Yes |
| 109 | HLA-B | NA | 54552 | -0.557 | 0.0007 | Yes |