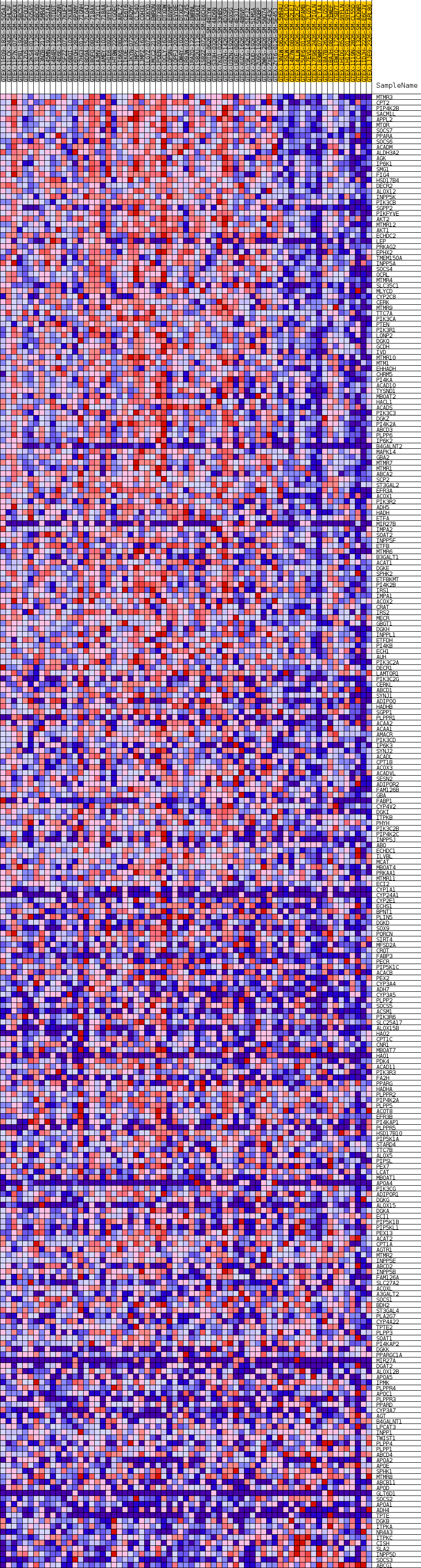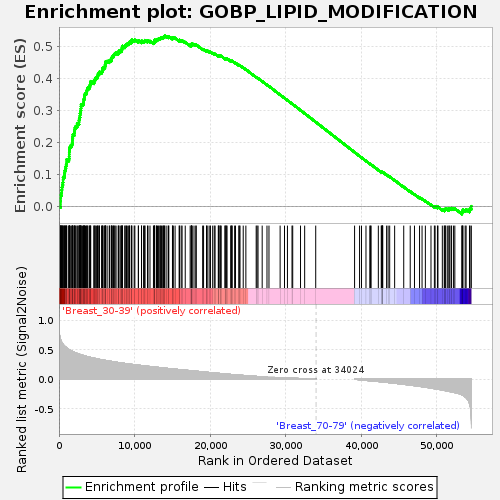
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Breast.Breast.cls #Breast_30-39_versus_Breast_70-79.Breast.cls #Breast_30-39_versus_Breast_70-79_repos |
| Phenotype | Breast.cls#Breast_30-39_versus_Breast_70-79_repos |
| Upregulated in class | Breast_30-39 |
| GeneSet | GOBP_LIPID_MODIFICATION |
| Enrichment Score (ES) | 0.53319573 |
| Normalized Enrichment Score (NES) | 1.7394637 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.486426 |
| FWER p-Value | 0.527 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | MTMR3 | NA | 209 | 0.668 | 0.0060 | Yes |
| 2 | CPT2 | NA | 210 | 0.668 | 0.0158 | Yes |
| 3 | PIP4K2B | NA | 221 | 0.663 | 0.0253 | Yes |
| 4 | SACM1L | NA | 227 | 0.660 | 0.0349 | Yes |
| 5 | APPL2 | NA | 333 | 0.629 | 0.0422 | Yes |
| 6 | MTOR | NA | 355 | 0.623 | 0.0510 | Yes |
| 7 | SOCS7 | NA | 386 | 0.616 | 0.0595 | Yes |
| 8 | PPARA | NA | 438 | 0.605 | 0.0674 | Yes |
| 9 | SOCS6 | NA | 515 | 0.591 | 0.0747 | Yes |
| 10 | ACADM | NA | 542 | 0.586 | 0.0828 | Yes |
| 11 | ALDH3A2 | NA | 550 | 0.585 | 0.0913 | Yes |
| 12 | AGK | NA | 698 | 0.562 | 0.0968 | Yes |
| 13 | IP6K1 | NA | 747 | 0.557 | 0.1041 | Yes |
| 14 | SMG1 | NA | 771 | 0.553 | 0.1118 | Yes |
| 15 | FIG4 | NA | 851 | 0.545 | 0.1184 | Yes |
| 16 | HSD17B4 | NA | 909 | 0.540 | 0.1253 | Yes |
| 17 | DECR2 | NA | 954 | 0.535 | 0.1323 | Yes |
| 18 | ALOX12 | NA | 1014 | 0.529 | 0.1390 | Yes |
| 19 | INPP5K | NA | 1017 | 0.528 | 0.1467 | Yes |
| 20 | PIK3CB | NA | 1307 | 0.502 | 0.1488 | Yes |
| 21 | SGPP2 | NA | 1343 | 0.499 | 0.1554 | Yes |
| 22 | PIKFYVE | NA | 1353 | 0.499 | 0.1626 | Yes |
| 23 | AKT2 | NA | 1358 | 0.498 | 0.1698 | Yes |
| 24 | MTMR12 | NA | 1368 | 0.497 | 0.1770 | Yes |
| 25 | AKT1 | NA | 1390 | 0.496 | 0.1839 | Yes |
| 26 | ECHDC2 | NA | 1496 | 0.489 | 0.1891 | Yes |
| 27 | LEP | NA | 1684 | 0.477 | 0.1927 | Yes |
| 28 | PRKAG2 | NA | 1741 | 0.473 | 0.1986 | Yes |
| 29 | EPHX2 | NA | 1777 | 0.469 | 0.2048 | Yes |
| 30 | TMEM150A | NA | 1783 | 0.469 | 0.2116 | Yes |
| 31 | INPP5A | NA | 1825 | 0.466 | 0.2177 | Yes |
| 32 | SOCS4 | NA | 1852 | 0.465 | 0.2240 | Yes |
| 33 | OCRL | NA | 2023 | 0.454 | 0.2276 | Yes |
| 34 | MTMR4 | NA | 2037 | 0.454 | 0.2340 | Yes |
| 35 | SLC35C1 | NA | 2077 | 0.452 | 0.2399 | Yes |
| 36 | MLYCD | NA | 2121 | 0.450 | 0.2457 | Yes |
| 37 | CYP2C8 | NA | 2215 | 0.445 | 0.2506 | Yes |
| 38 | CERK | NA | 2395 | 0.436 | 0.2537 | Yes |
| 39 | MTMR9 | NA | 2410 | 0.435 | 0.2598 | Yes |
| 40 | TTC7A | NA | 2647 | 0.425 | 0.2617 | Yes |
| 41 | PIK3CA | NA | 2652 | 0.424 | 0.2678 | Yes |
| 42 | PTEN | NA | 2673 | 0.423 | 0.2737 | Yes |
| 43 | PIK3R1 | NA | 2739 | 0.420 | 0.2786 | Yes |
| 44 | LONP2 | NA | 2778 | 0.419 | 0.2841 | Yes |
| 45 | DGKQ | NA | 2788 | 0.418 | 0.2901 | Yes |
| 46 | GCDH | NA | 2826 | 0.416 | 0.2955 | Yes |
| 47 | IVD | NA | 2878 | 0.414 | 0.3006 | Yes |
| 48 | MTMR10 | NA | 2904 | 0.413 | 0.3062 | Yes |
| 49 | MTM1 | NA | 2916 | 0.412 | 0.3121 | Yes |
| 50 | EHHADH | NA | 2929 | 0.412 | 0.3179 | Yes |
| 51 | CHRM5 | NA | 3119 | 0.405 | 0.3204 | Yes |
| 52 | PI4KA | NA | 3246 | 0.400 | 0.3239 | Yes |
| 53 | ACAD10 | NA | 3267 | 0.399 | 0.3294 | Yes |
| 54 | TYSND1 | NA | 3274 | 0.399 | 0.3352 | Yes |
| 55 | MBOAT2 | NA | 3362 | 0.396 | 0.3394 | Yes |
| 56 | HACL1 | NA | 3370 | 0.396 | 0.3451 | Yes |
| 57 | ACADS | NA | 3399 | 0.395 | 0.3503 | Yes |
| 58 | PIK3C3 | NA | 3507 | 0.391 | 0.3541 | Yes |
| 59 | DGKZ | NA | 3668 | 0.385 | 0.3568 | Yes |
| 60 | PI4K2A | NA | 3675 | 0.385 | 0.3623 | Yes |
| 61 | ABCD3 | NA | 3733 | 0.382 | 0.3669 | Yes |
| 62 | PLPP6 | NA | 3812 | 0.379 | 0.3710 | Yes |
| 63 | IP6K2 | NA | 4009 | 0.374 | 0.3729 | Yes |
| 64 | B4GALNT2 | NA | 4082 | 0.372 | 0.3770 | Yes |
| 65 | MAPK14 | NA | 4118 | 0.371 | 0.3818 | Yes |
| 66 | GBA2 | NA | 4144 | 0.370 | 0.3868 | Yes |
| 67 | MTMR7 | NA | 4199 | 0.368 | 0.3912 | Yes |
| 68 | MTMR1 | NA | 4630 | 0.356 | 0.3885 | Yes |
| 69 | ABCA2 | NA | 4634 | 0.355 | 0.3937 | Yes |
| 70 | SCP2 | NA | 4749 | 0.352 | 0.3968 | Yes |
| 71 | ST3GAL2 | NA | 4793 | 0.351 | 0.4011 | Yes |
| 72 | EFR3A | NA | 4949 | 0.347 | 0.4034 | Yes |
| 73 | ACOX1 | NA | 5098 | 0.343 | 0.4057 | Yes |
| 74 | PIK3R2 | NA | 5143 | 0.342 | 0.4099 | Yes |
| 75 | ADH5 | NA | 5170 | 0.341 | 0.4144 | Yes |
| 76 | HADH | NA | 5324 | 0.336 | 0.4165 | Yes |
| 77 | ETFA | NA | 5356 | 0.336 | 0.4209 | Yes |
| 78 | MIR27B | NA | 5640 | 0.329 | 0.4205 | Yes |
| 79 | IMPA2 | NA | 5708 | 0.327 | 0.4241 | Yes |
| 80 | SOAT2 | NA | 5744 | 0.326 | 0.4282 | Yes |
| 81 | INPP5F | NA | 5763 | 0.326 | 0.4327 | Yes |
| 82 | ETFB | NA | 5943 | 0.322 | 0.4341 | Yes |
| 83 | MTMR6 | NA | 6066 | 0.319 | 0.4365 | Yes |
| 84 | B3GALT1 | NA | 6101 | 0.318 | 0.4406 | Yes |
| 85 | ACAT1 | NA | 6111 | 0.318 | 0.4451 | Yes |
| 86 | DGKE | NA | 6163 | 0.317 | 0.4488 | Yes |
| 87 | SPHK2 | NA | 6175 | 0.317 | 0.4532 | Yes |
| 88 | ETFBKMT | NA | 6372 | 0.312 | 0.4542 | Yes |
| 89 | PI4K2B | NA | 6648 | 0.306 | 0.4537 | Yes |
| 90 | IRS1 | NA | 6700 | 0.305 | 0.4572 | Yes |
| 91 | IMPA1 | NA | 6915 | 0.301 | 0.4577 | Yes |
| 92 | ACOX2 | NA | 6955 | 0.300 | 0.4614 | Yes |
| 93 | CRAT | NA | 6969 | 0.300 | 0.4655 | Yes |
| 94 | IRS2 | NA | 7052 | 0.298 | 0.4684 | Yes |
| 95 | MECR | NA | 7192 | 0.295 | 0.4702 | Yes |
| 96 | GBGT1 | NA | 7235 | 0.295 | 0.4737 | Yes |
| 97 | DGKH | NA | 7314 | 0.293 | 0.4766 | Yes |
| 98 | INPPL1 | NA | 7421 | 0.290 | 0.4789 | Yes |
| 99 | ETFDH | NA | 7555 | 0.288 | 0.4807 | Yes |
| 100 | PI4KB | NA | 7834 | 0.282 | 0.4797 | Yes |
| 101 | ECH1 | NA | 7889 | 0.281 | 0.4828 | Yes |
| 102 | AUH | NA | 7930 | 0.280 | 0.4862 | Yes |
| 103 | PIK3C2A | NA | 8112 | 0.277 | 0.4869 | Yes |
| 104 | DECR1 | NA | 8300 | 0.274 | 0.4875 | Yes |
| 105 | LAMTOR1 | NA | 8320 | 0.273 | 0.4912 | Yes |
| 106 | PIK3C2G | NA | 8342 | 0.273 | 0.4948 | Yes |
| 107 | CERKL | NA | 8354 | 0.273 | 0.4986 | Yes |
| 108 | ABCD1 | NA | 8410 | 0.272 | 0.5016 | Yes |
| 109 | SYNJ1 | NA | 8722 | 0.266 | 0.4997 | Yes |
| 110 | ADIPOQ | NA | 8737 | 0.266 | 0.5034 | Yes |
| 111 | HADHB | NA | 8884 | 0.263 | 0.5046 | Yes |
| 112 | SGPP1 | NA | 8935 | 0.262 | 0.5075 | Yes |
| 113 | PLPPR1 | NA | 9099 | 0.259 | 0.5083 | Yes |
| 114 | ACAA2 | NA | 9231 | 0.257 | 0.5097 | Yes |
| 115 | ACAA1 | NA | 9324 | 0.255 | 0.5117 | Yes |
| 116 | AMACR | NA | 9352 | 0.255 | 0.5150 | Yes |
| 117 | PIK3CD | NA | 9579 | 0.251 | 0.5145 | Yes |
| 118 | IP6K3 | NA | 9615 | 0.251 | 0.5175 | Yes |
| 119 | SYNJ2 | NA | 9642 | 0.250 | 0.5207 | Yes |
| 120 | ACADL | NA | 9951 | 0.245 | 0.5187 | Yes |
| 121 | CPT1B | NA | 10043 | 0.244 | 0.5206 | Yes |
| 122 | ACOX3 | NA | 10503 | 0.236 | 0.5156 | Yes |
| 123 | ACADVL | NA | 10516 | 0.236 | 0.5188 | Yes |
| 124 | SESN2 | NA | 10921 | 0.230 | 0.5148 | Yes |
| 125 | ADIPOR2 | NA | 10925 | 0.230 | 0.5181 | Yes |
| 126 | FAM126B | NA | 11198 | 0.226 | 0.5164 | Yes |
| 127 | GBA | NA | 11336 | 0.224 | 0.5172 | Yes |
| 128 | FABP1 | NA | 11383 | 0.223 | 0.5196 | Yes |
| 129 | CYP4V2 | NA | 11718 | 0.218 | 0.5167 | Yes |
| 130 | DGKI | NA | 11780 | 0.217 | 0.5187 | Yes |
| 131 | ITPKB | NA | 12046 | 0.214 | 0.5170 | Yes |
| 132 | PHYH | NA | 12503 | 0.208 | 0.5116 | Yes |
| 133 | PIK3C2B | NA | 12594 | 0.206 | 0.5130 | Yes |
| 134 | PIP4K2C | NA | 12621 | 0.206 | 0.5156 | Yes |
| 135 | INPP5J | NA | 12632 | 0.206 | 0.5184 | Yes |
| 136 | ABO | NA | 12675 | 0.206 | 0.5207 | Yes |
| 137 | ECHDC1 | NA | 12905 | 0.202 | 0.5194 | Yes |
| 138 | ILVBL | NA | 12916 | 0.202 | 0.5222 | Yes |
| 139 | MCAT | NA | 13040 | 0.200 | 0.5229 | Yes |
| 140 | MBOAT4 | NA | 13156 | 0.199 | 0.5237 | Yes |
| 141 | PRKAA1 | NA | 13261 | 0.197 | 0.5246 | Yes |
| 142 | MTMR11 | NA | 13399 | 0.195 | 0.5250 | Yes |
| 143 | ECI2 | NA | 13465 | 0.194 | 0.5266 | Yes |
| 144 | CYP1A1 | NA | 13576 | 0.192 | 0.5274 | Yes |
| 145 | CYP24A1 | NA | 13736 | 0.190 | 0.5273 | Yes |
| 146 | CYP2E1 | NA | 13816 | 0.189 | 0.5286 | Yes |
| 147 | ECHS1 | NA | 13864 | 0.189 | 0.5305 | Yes |
| 148 | BPNT1 | NA | 13975 | 0.187 | 0.5312 | Yes |
| 149 | PLIN5 | NA | 14018 | 0.186 | 0.5332 | Yes |
| 150 | DGKD | NA | 14282 | 0.183 | 0.5310 | No |
| 151 | SOX9 | NA | 14498 | 0.180 | 0.5297 | No |
| 152 | PORCN | NA | 14535 | 0.180 | 0.5317 | No |
| 153 | SIRT4 | NA | 15022 | 0.173 | 0.5253 | No |
| 154 | MFSD2A | NA | 15023 | 0.173 | 0.5278 | No |
| 155 | CROT | NA | 15134 | 0.172 | 0.5283 | No |
| 156 | FABP3 | NA | 15369 | 0.169 | 0.5265 | No |
| 157 | PECR | NA | 15908 | 0.162 | 0.5190 | No |
| 158 | PIP5K1C | NA | 15944 | 0.162 | 0.5207 | No |
| 159 | ACACB | NA | 16170 | 0.159 | 0.5189 | No |
| 160 | PEX2 | NA | 16293 | 0.158 | 0.5190 | No |
| 161 | CYP3A4 | NA | 16711 | 0.153 | 0.5136 | No |
| 162 | ADH7 | NA | 17365 | 0.145 | 0.5037 | No |
| 163 | CYP3A5 | NA | 17513 | 0.143 | 0.5031 | No |
| 164 | PLPP2 | NA | 17538 | 0.143 | 0.5047 | No |
| 165 | SOCS5 | NA | 17545 | 0.143 | 0.5067 | No |
| 166 | ACSM1 | NA | 17550 | 0.142 | 0.5087 | No |
| 167 | PIK3R6 | NA | 17640 | 0.141 | 0.5092 | No |
| 168 | SLC25A17 | NA | 17872 | 0.138 | 0.5069 | No |
| 169 | ALOX15B | NA | 18108 | 0.135 | 0.5046 | No |
| 170 | HAO2 | NA | 18169 | 0.135 | 0.5055 | No |
| 171 | CPT1C | NA | 19017 | 0.124 | 0.4917 | No |
| 172 | CNR1 | NA | 19137 | 0.122 | 0.4913 | No |
| 173 | MBOAT7 | NA | 19526 | 0.117 | 0.4859 | No |
| 174 | HAO1 | NA | 19558 | 0.117 | 0.4870 | No |
| 175 | PDK4 | NA | 19710 | 0.115 | 0.4859 | No |
| 176 | ACAD11 | NA | 19969 | 0.112 | 0.4828 | No |
| 177 | PIK3R3 | NA | 20047 | 0.111 | 0.4830 | No |
| 178 | FA2H | NA | 20344 | 0.107 | 0.4791 | No |
| 179 | PPARG | NA | 20553 | 0.105 | 0.4768 | No |
| 180 | HADHA | NA | 20662 | 0.104 | 0.4764 | No |
| 181 | PLPPR2 | NA | 21087 | 0.098 | 0.4700 | No |
| 182 | PIP4K2A | NA | 21146 | 0.098 | 0.4704 | No |
| 183 | PLPP5 | NA | 21150 | 0.098 | 0.4718 | No |
| 184 | ACOT8 | NA | 21338 | 0.096 | 0.4697 | No |
| 185 | EFR3B | NA | 21342 | 0.096 | 0.4711 | No |
| 186 | PI4KAP1 | NA | 21470 | 0.094 | 0.4701 | No |
| 187 | PLPPR5 | NA | 21981 | 0.088 | 0.4620 | No |
| 188 | HSD17B10 | NA | 21987 | 0.088 | 0.4632 | No |
| 189 | PIP5K1A | NA | 22104 | 0.087 | 0.4624 | No |
| 190 | STARD4 | NA | 22145 | 0.087 | 0.4629 | No |
| 191 | TTC7B | NA | 22246 | 0.086 | 0.4623 | No |
| 192 | ALOX5 | NA | 22730 | 0.080 | 0.4546 | No |
| 193 | PIPSL | NA | 22816 | 0.079 | 0.4542 | No |
| 194 | PEX7 | NA | 22878 | 0.079 | 0.4542 | No |
| 195 | LCAT | NA | 22918 | 0.078 | 0.4547 | No |
| 196 | MBOAT1 | NA | 23251 | 0.075 | 0.4497 | No |
| 197 | APOA4 | NA | 23367 | 0.073 | 0.4486 | No |
| 198 | PIK3CG | NA | 23809 | 0.069 | 0.4415 | No |
| 199 | ADIPOR1 | NA | 23859 | 0.068 | 0.4416 | No |
| 200 | DGKG | NA | 23961 | 0.067 | 0.4407 | No |
| 201 | ALOX15 | NA | 24387 | 0.063 | 0.4338 | No |
| 202 | DGKA | NA | 24737 | 0.060 | 0.4283 | No |
| 203 | ECI1 | NA | 26103 | 0.047 | 0.4039 | No |
| 204 | PIP5K1B | NA | 26213 | 0.046 | 0.4025 | No |
| 205 | PIP5KL1 | NA | 26347 | 0.045 | 0.4007 | No |
| 206 | PEX13 | NA | 26897 | 0.040 | 0.3912 | No |
| 207 | ACAT2 | NA | 27532 | 0.036 | 0.3801 | No |
| 208 | CPT1A | NA | 27796 | 0.034 | 0.3757 | No |
| 209 | AGTR1 | NA | 29272 | 0.024 | 0.3489 | No |
| 210 | MTMR2 | NA | 29851 | 0.021 | 0.3386 | No |
| 211 | INPP5E | NA | 30241 | 0.019 | 0.3317 | No |
| 212 | ABCD2 | NA | 30846 | 0.016 | 0.3208 | No |
| 213 | INPP5B | NA | 30940 | 0.016 | 0.3194 | No |
| 214 | FAM126A | NA | 31972 | 0.011 | 0.3005 | No |
| 215 | SLC27A2 | NA | 32521 | 0.009 | 0.2906 | No |
| 216 | ACOXL | NA | 33984 | 0.001 | 0.2637 | No |
| 217 | A3GALT2 | NA | 39129 | -0.000 | 0.1690 | No |
| 218 | SOCS1 | NA | 39789 | -0.009 | 0.1570 | No |
| 219 | BDH2 | NA | 40047 | -0.012 | 0.1524 | No |
| 220 | ST3GAL4 | NA | 40638 | -0.019 | 0.1419 | No |
| 221 | PLA2G7 | NA | 41144 | -0.025 | 0.1329 | No |
| 222 | CYP4A22 | NA | 41245 | -0.027 | 0.1315 | No |
| 223 | TPTE2 | NA | 41312 | -0.027 | 0.1307 | No |
| 224 | PLPP3 | NA | 42273 | -0.039 | 0.1136 | No |
| 225 | SOAT1 | NA | 42659 | -0.044 | 0.1071 | No |
| 226 | PI4KAP2 | NA | 42713 | -0.045 | 0.1068 | No |
| 227 | DGKK | NA | 42791 | -0.046 | 0.1061 | No |
| 228 | PPARGC1A | NA | 42813 | -0.046 | 0.1064 | No |
| 229 | MIR27A | NA | 42819 | -0.046 | 0.1069 | No |
| 230 | DGAT2 | NA | 42859 | -0.047 | 0.1069 | No |
| 231 | ALOX12B | NA | 43369 | -0.053 | 0.0983 | No |
| 232 | APOA5 | NA | 43566 | -0.056 | 0.0955 | No |
| 233 | IPMK | NA | 43777 | -0.059 | 0.0925 | No |
| 234 | PLPPR4 | NA | 44432 | -0.068 | 0.0815 | No |
| 235 | APOC1 | NA | 45635 | -0.086 | 0.0606 | No |
| 236 | PLPPR3 | NA | 46494 | -0.099 | 0.0463 | No |
| 237 | PPARD | NA | 47058 | -0.108 | 0.0375 | No |
| 238 | CYP3A7 | NA | 47729 | -0.120 | 0.0270 | No |
| 239 | AGT | NA | 48061 | -0.127 | 0.0227 | No |
| 240 | B4GALNT1 | NA | 48499 | -0.135 | 0.0167 | No |
| 241 | LPCAT3 | NA | 49243 | -0.150 | 0.0052 | No |
| 242 | INPP1 | NA | 49692 | -0.159 | -0.0007 | No |
| 243 | TWIST1 | NA | 49789 | -0.162 | -0.0001 | No |
| 244 | PLPP4 | NA | 50099 | -0.169 | -0.0033 | No |
| 245 | PLPP1 | NA | 50166 | -0.170 | -0.0020 | No |
| 246 | ABCD4 | NA | 50744 | -0.183 | -0.0100 | No |
| 247 | APOA2 | NA | 50997 | -0.189 | -0.0118 | No |
| 248 | APOE | NA | 51066 | -0.191 | -0.0103 | No |
| 249 | SPHK1 | NA | 51119 | -0.192 | -0.0084 | No |
| 250 | MTMR8 | NA | 51205 | -0.194 | -0.0071 | No |
| 251 | ABCB11 | NA | 51458 | -0.201 | -0.0088 | No |
| 252 | APOD | NA | 51633 | -0.206 | -0.0090 | No |
| 253 | GLT6D1 | NA | 51655 | -0.207 | -0.0064 | No |
| 254 | SOCS2 | NA | 51897 | -0.212 | -0.0077 | No |
| 255 | APOA1 | NA | 51971 | -0.214 | -0.0059 | No |
| 256 | ADH4 | NA | 52234 | -0.221 | -0.0075 | No |
| 257 | TPTE | NA | 52400 | -0.226 | -0.0072 | No |
| 258 | DGKB | NA | 53337 | -0.263 | -0.0206 | No |
| 259 | ITPKA | NA | 53363 | -0.265 | -0.0171 | No |
| 260 | NR4A3 | NA | 53426 | -0.270 | -0.0143 | No |
| 261 | ITPKC | NA | 53473 | -0.273 | -0.0111 | No |
| 262 | CISH | NA | 53769 | -0.299 | -0.0122 | No |
| 263 | SLA2 | NA | 53892 | -0.311 | -0.0099 | No |
| 264 | INPP5D | NA | 54339 | -0.407 | -0.0121 | No |
| 265 | SOCS3 | NA | 54415 | -0.440 | -0.0070 | No |
| 266 | ABCG1 | NA | 54585 | -0.697 | 0.0001 | No |