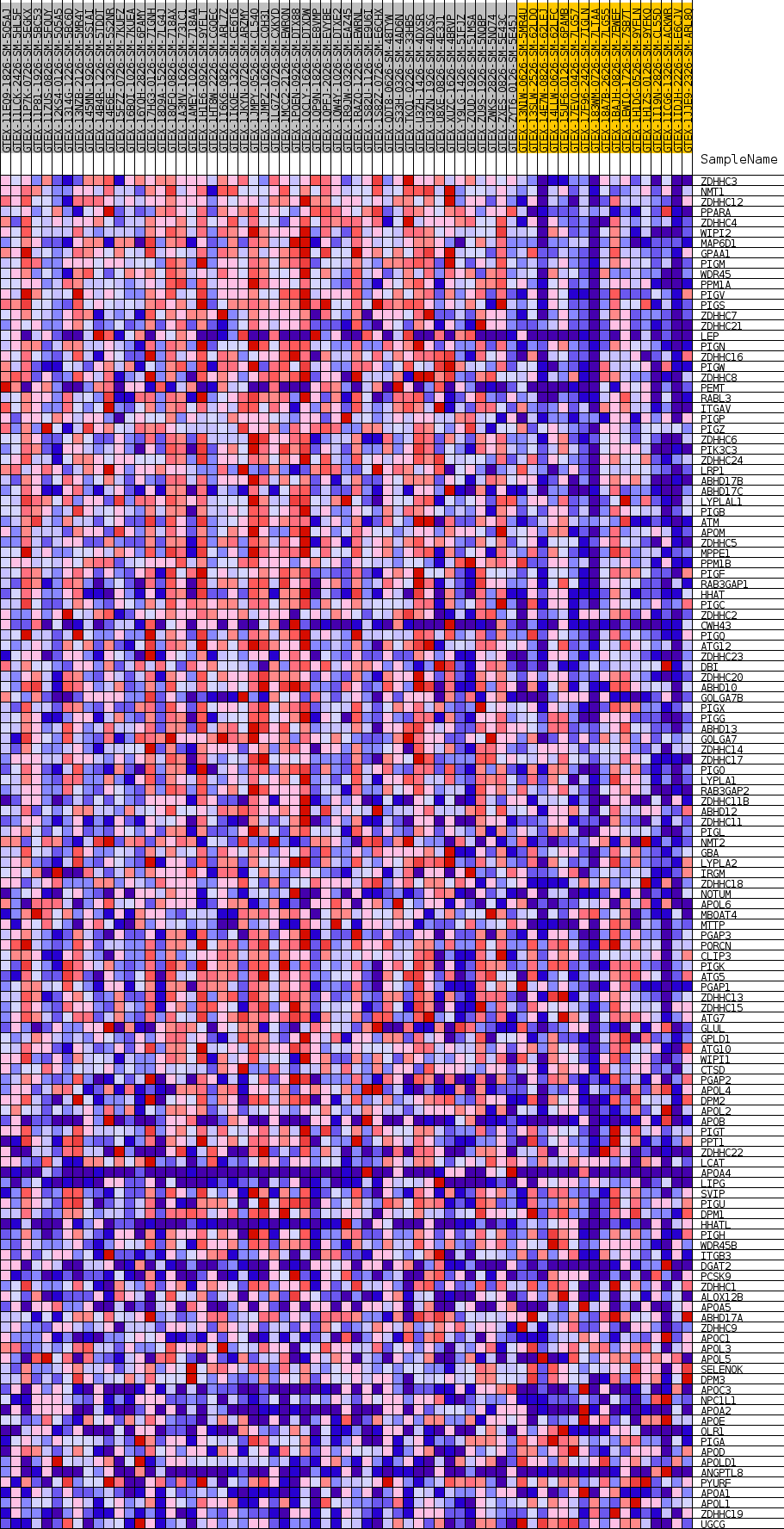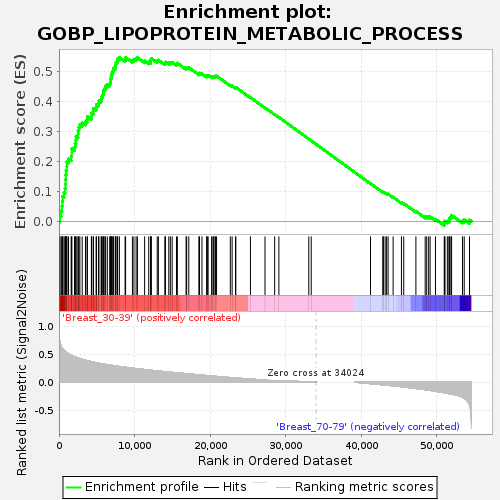
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Breast.Breast.cls #Breast_30-39_versus_Breast_70-79.Breast.cls #Breast_30-39_versus_Breast_70-79_repos |
| Phenotype | Breast.cls#Breast_30-39_versus_Breast_70-79_repos |
| Upregulated in class | Breast_30-39 |
| GeneSet | GOBP_LIPOPROTEIN_METABOLIC_PROCESS |
| Enrichment Score (ES) | 0.5481111 |
| Normalized Enrichment Score (NES) | 1.6620649 |
| Nominal p-value | 0.0068728523 |
| FDR q-value | 0.24203251 |
| FWER p-Value | 0.768 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | ZDHHC3 | NA | 116 | 0.718 | 0.0186 | Yes |
| 2 | NMT1 | NA | 276 | 0.648 | 0.0344 | Yes |
| 3 | ZDHHC12 | NA | 385 | 0.617 | 0.0502 | Yes |
| 4 | PPARA | NA | 438 | 0.605 | 0.0667 | Yes |
| 5 | ZDHHC4 | NA | 478 | 0.598 | 0.0832 | Yes |
| 6 | WIPI2 | NA | 656 | 0.569 | 0.0964 | Yes |
| 7 | MAP6D1 | NA | 787 | 0.552 | 0.1099 | Yes |
| 8 | GPAA1 | NA | 854 | 0.545 | 0.1244 | Yes |
| 9 | PIGM | NA | 877 | 0.543 | 0.1397 | Yes |
| 10 | WDR45 | NA | 905 | 0.541 | 0.1548 | Yes |
| 11 | PPM1A | NA | 935 | 0.536 | 0.1698 | Yes |
| 12 | PIGV | NA | 1029 | 0.528 | 0.1833 | Yes |
| 13 | PIGS | NA | 1039 | 0.526 | 0.1983 | Yes |
| 14 | ZDHHC7 | NA | 1258 | 0.507 | 0.2089 | Yes |
| 15 | ZDHHC21 | NA | 1609 | 0.482 | 0.2164 | Yes |
| 16 | LEP | NA | 1684 | 0.477 | 0.2288 | Yes |
| 17 | PIGN | NA | 1685 | 0.477 | 0.2426 | Yes |
| 18 | ZDHHC16 | NA | 2052 | 0.453 | 0.2489 | Yes |
| 19 | PIGW | NA | 2130 | 0.449 | 0.2605 | Yes |
| 20 | ZDHHC8 | NA | 2231 | 0.444 | 0.2714 | Yes |
| 21 | PEMT | NA | 2261 | 0.442 | 0.2837 | Yes |
| 22 | RABL3 | NA | 2523 | 0.430 | 0.2913 | Yes |
| 23 | ITGAV | NA | 2531 | 0.430 | 0.3036 | Yes |
| 24 | PIGP | NA | 2621 | 0.426 | 0.3142 | Yes |
| 25 | PIGZ | NA | 2762 | 0.419 | 0.3238 | Yes |
| 26 | ZDHHC6 | NA | 3072 | 0.406 | 0.3298 | Yes |
| 27 | PIK3C3 | NA | 3507 | 0.391 | 0.3331 | Yes |
| 28 | ZDHHC24 | NA | 3715 | 0.383 | 0.3404 | Yes |
| 29 | LRP1 | NA | 3758 | 0.382 | 0.3506 | Yes |
| 30 | ABHD17B | NA | 4287 | 0.366 | 0.3515 | Yes |
| 31 | ABHD17C | NA | 4308 | 0.365 | 0.3616 | Yes |
| 32 | LYPLAL1 | NA | 4533 | 0.358 | 0.3679 | Yes |
| 33 | PIGB | NA | 4552 | 0.358 | 0.3779 | Yes |
| 34 | ATM | NA | 4915 | 0.348 | 0.3813 | Yes |
| 35 | APOM | NA | 4957 | 0.347 | 0.3905 | Yes |
| 36 | ZDHHC5 | NA | 5224 | 0.339 | 0.3954 | Yes |
| 37 | MPPE1 | NA | 5286 | 0.338 | 0.4040 | Yes |
| 38 | PPM1B | NA | 5573 | 0.331 | 0.4083 | Yes |
| 39 | PIGF | NA | 5633 | 0.329 | 0.4167 | Yes |
| 40 | RAB3GAP1 | NA | 5790 | 0.325 | 0.4233 | Yes |
| 41 | HHAT | NA | 5870 | 0.323 | 0.4311 | Yes |
| 42 | PIGC | NA | 5925 | 0.322 | 0.4394 | Yes |
| 43 | ZDHHC2 | NA | 6079 | 0.319 | 0.4458 | Yes |
| 44 | CWH43 | NA | 6171 | 0.317 | 0.4533 | Yes |
| 45 | PIGQ | NA | 6397 | 0.312 | 0.4582 | Yes |
| 46 | ATG12 | NA | 6712 | 0.305 | 0.4612 | Yes |
| 47 | ZDHHC23 | NA | 6827 | 0.303 | 0.4679 | Yes |
| 48 | DBI | NA | 6844 | 0.302 | 0.4763 | Yes |
| 49 | ZDHHC20 | NA | 6894 | 0.301 | 0.4841 | Yes |
| 50 | ABHD10 | NA | 6962 | 0.300 | 0.4915 | Yes |
| 51 | GOLGA7B | NA | 7041 | 0.298 | 0.4987 | Yes |
| 52 | PIGX | NA | 7148 | 0.296 | 0.5053 | Yes |
| 53 | PIGG | NA | 7224 | 0.295 | 0.5124 | Yes |
| 54 | ABHD13 | NA | 7450 | 0.290 | 0.5166 | Yes |
| 55 | GOLGA7 | NA | 7487 | 0.289 | 0.5243 | Yes |
| 56 | ZDHHC14 | NA | 7552 | 0.288 | 0.5315 | Yes |
| 57 | ZDHHC17 | NA | 7732 | 0.284 | 0.5364 | Yes |
| 58 | PIGO | NA | 7764 | 0.284 | 0.5440 | Yes |
| 59 | LYPLA1 | NA | 7994 | 0.279 | 0.5478 | Yes |
| 60 | RAB3GAP2 | NA | 8749 | 0.266 | 0.5417 | Yes |
| 61 | ZDHHC11B | NA | 8815 | 0.265 | 0.5481 | Yes |
| 62 | ABHD12 | NA | 9720 | 0.249 | 0.5387 | No |
| 63 | ZDHHC11 | NA | 9920 | 0.246 | 0.5421 | No |
| 64 | PIGL | NA | 10207 | 0.241 | 0.5438 | No |
| 65 | NMT2 | NA | 10381 | 0.238 | 0.5475 | No |
| 66 | GBA | NA | 11336 | 0.224 | 0.5365 | No |
| 67 | LYPLA2 | NA | 11880 | 0.216 | 0.5328 | No |
| 68 | IRGM | NA | 12128 | 0.213 | 0.5344 | No |
| 69 | ZDHHC18 | NA | 12142 | 0.213 | 0.5403 | No |
| 70 | NOTUM | NA | 12226 | 0.212 | 0.5449 | No |
| 71 | APOL6 | NA | 13003 | 0.201 | 0.5364 | No |
| 72 | MBOAT4 | NA | 13156 | 0.199 | 0.5393 | No |
| 73 | MTTP | NA | 14012 | 0.186 | 0.5290 | No |
| 74 | PGAP3 | NA | 14096 | 0.185 | 0.5328 | No |
| 75 | PORCN | NA | 14535 | 0.180 | 0.5300 | No |
| 76 | CLIP3 | NA | 14738 | 0.177 | 0.5314 | No |
| 77 | PIGK | NA | 14980 | 0.174 | 0.5320 | No |
| 78 | ATG5 | NA | 15552 | 0.167 | 0.5263 | No |
| 79 | PGAP1 | NA | 15667 | 0.166 | 0.5290 | No |
| 80 | ZDHHC13 | NA | 16823 | 0.151 | 0.5122 | No |
| 81 | ZDHHC15 | NA | 16900 | 0.150 | 0.5151 | No |
| 82 | ATG7 | NA | 17182 | 0.147 | 0.5142 | No |
| 83 | GLUL | NA | 18517 | 0.130 | 0.4934 | No |
| 84 | GPLD1 | NA | 18598 | 0.129 | 0.4957 | No |
| 85 | ATG10 | NA | 18926 | 0.125 | 0.4933 | No |
| 86 | WIPI1 | NA | 19536 | 0.117 | 0.4855 | No |
| 87 | CTSD | NA | 19641 | 0.116 | 0.4869 | No |
| 88 | PGAP2 | NA | 19740 | 0.114 | 0.4884 | No |
| 89 | APOL4 | NA | 20200 | 0.109 | 0.4831 | No |
| 90 | DPM2 | NA | 20379 | 0.107 | 0.4829 | No |
| 91 | APOL2 | NA | 20469 | 0.106 | 0.4844 | No |
| 92 | APOB | NA | 20653 | 0.104 | 0.4840 | No |
| 93 | PIGT | NA | 20709 | 0.103 | 0.4860 | No |
| 94 | PPT1 | NA | 20855 | 0.101 | 0.4862 | No |
| 95 | ZDHHC22 | NA | 22678 | 0.081 | 0.4551 | No |
| 96 | LCAT | NA | 22918 | 0.078 | 0.4530 | No |
| 97 | APOA4 | NA | 23367 | 0.073 | 0.4468 | No |
| 98 | LIPG | NA | 23410 | 0.073 | 0.4482 | No |
| 99 | SVIP | NA | 25343 | 0.054 | 0.4143 | No |
| 100 | PIGU | NA | 27270 | 0.037 | 0.3800 | No |
| 101 | DPM1 | NA | 28550 | 0.029 | 0.3573 | No |
| 102 | HHATL | NA | 29108 | 0.026 | 0.3478 | No |
| 103 | PIGH | NA | 33069 | 0.007 | 0.2753 | No |
| 104 | WDR45B | NA | 33389 | 0.005 | 0.2696 | No |
| 105 | ITGB3 | NA | 41238 | -0.026 | 0.1263 | No |
| 106 | DGAT2 | NA | 42859 | -0.047 | 0.0979 | No |
| 107 | PCSK9 | NA | 42943 | -0.048 | 0.0977 | No |
| 108 | ZDHHC1 | NA | 43172 | -0.051 | 0.0950 | No |
| 109 | ALOX12B | NA | 43369 | -0.053 | 0.0929 | No |
| 110 | APOA5 | NA | 43566 | -0.056 | 0.0909 | No |
| 111 | ABHD17A | NA | 44231 | -0.065 | 0.0806 | No |
| 112 | ZDHHC9 | NA | 45333 | -0.081 | 0.0628 | No |
| 113 | APOC1 | NA | 45635 | -0.086 | 0.0597 | No |
| 114 | APOL3 | NA | 47246 | -0.112 | 0.0334 | No |
| 115 | APOL5 | NA | 48474 | -0.134 | 0.0147 | No |
| 116 | SELENOK | NA | 48675 | -0.138 | 0.0150 | No |
| 117 | DPM3 | NA | 48935 | -0.143 | 0.0144 | No |
| 118 | APOC3 | NA | 49129 | -0.147 | 0.0151 | No |
| 119 | NPC1L1 | NA | 49838 | -0.163 | 0.0068 | No |
| 120 | APOA2 | NA | 50997 | -0.189 | -0.0090 | No |
| 121 | APOE | NA | 51066 | -0.191 | -0.0047 | No |
| 122 | OLR1 | NA | 51075 | -0.191 | 0.0006 | No |
| 123 | PIGA | NA | 51410 | -0.200 | 0.0003 | No |
| 124 | APOD | NA | 51633 | -0.206 | 0.0021 | No |
| 125 | APOLD1 | NA | 51647 | -0.206 | 0.0078 | No |
| 126 | ANGPTL8 | NA | 51737 | -0.208 | 0.0122 | No |
| 127 | PYURF | NA | 51900 | -0.212 | 0.0154 | No |
| 128 | APOA1 | NA | 51971 | -0.214 | 0.0203 | No |
| 129 | APOL1 | NA | 53412 | -0.268 | 0.0016 | No |
| 130 | ZDHHC19 | NA | 53642 | -0.286 | 0.0056 | No |
| 131 | UGCG | NA | 54345 | -0.409 | 0.0045 | No |