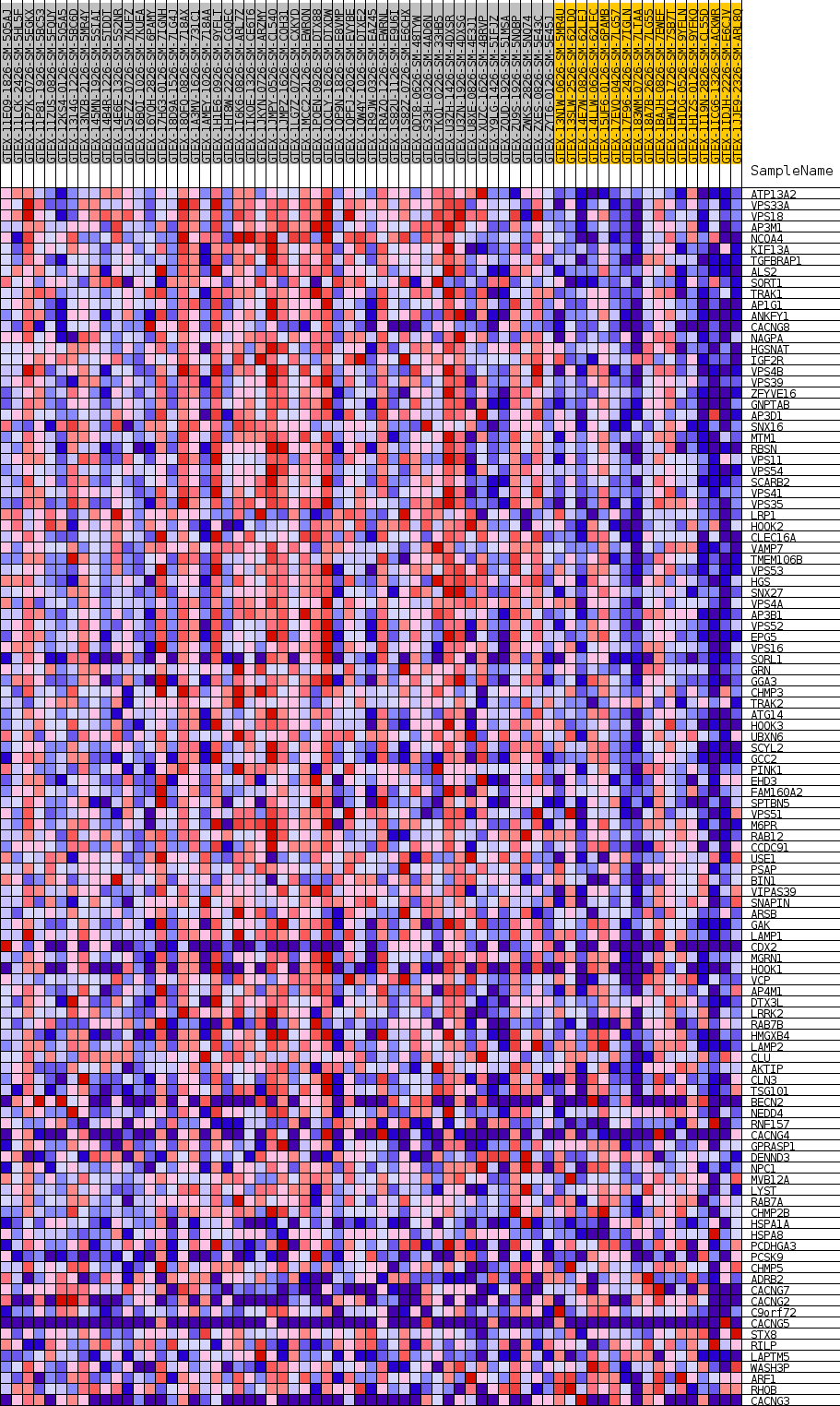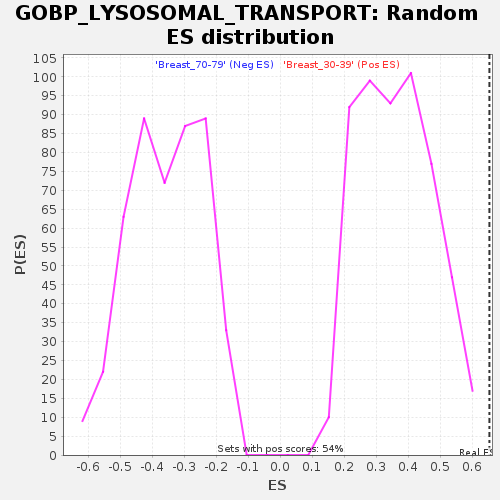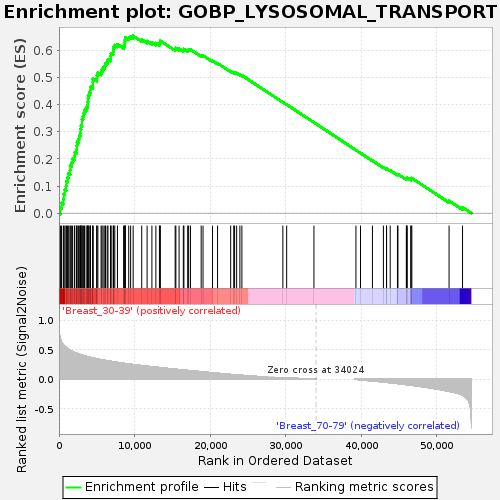
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Breast.Breast.cls #Breast_30-39_versus_Breast_70-79.Breast.cls #Breast_30-39_versus_Breast_70-79_repos |
| Phenotype | Breast.cls#Breast_30-39_versus_Breast_70-79_repos |
| Upregulated in class | Breast_30-39 |
| GeneSet | GOBP_LYSOSOMAL_TRANSPORT |
| Enrichment Score (ES) | 0.65284854 |
| Normalized Enrichment Score (NES) | 1.8071036 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.8077459 |
| FWER p-Value | 0.32 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | ATP13A2 | NA | 188 | 0.681 | 0.0190 | Yes |
| 2 | VPS33A | NA | 335 | 0.629 | 0.0371 | Yes |
| 3 | VPS18 | NA | 571 | 0.582 | 0.0520 | Yes |
| 4 | AP3M1 | NA | 623 | 0.573 | 0.0700 | Yes |
| 5 | NCOA4 | NA | 773 | 0.553 | 0.0855 | Yes |
| 6 | KIF13A | NA | 942 | 0.536 | 0.1001 | Yes |
| 7 | TGFBRAP1 | NA | 976 | 0.532 | 0.1171 | Yes |
| 8 | ALS2 | NA | 1136 | 0.518 | 0.1313 | Yes |
| 9 | SORT1 | NA | 1238 | 0.509 | 0.1462 | Yes |
| 10 | TRAK1 | NA | 1465 | 0.491 | 0.1583 | Yes |
| 11 | AP1G1 | NA | 1483 | 0.489 | 0.1741 | Yes |
| 12 | ANKFY1 | NA | 1678 | 0.477 | 0.1863 | Yes |
| 13 | CACNG8 | NA | 1776 | 0.469 | 0.2000 | Yes |
| 14 | NAGPA | NA | 2030 | 0.454 | 0.2104 | Yes |
| 15 | HGSNAT | NA | 2086 | 0.451 | 0.2243 | Yes |
| 16 | IGF2R | NA | 2327 | 0.439 | 0.2344 | Yes |
| 17 | VPS4B | NA | 2362 | 0.438 | 0.2482 | Yes |
| 18 | VPS39 | NA | 2386 | 0.436 | 0.2622 | Yes |
| 19 | ZFYVE16 | NA | 2596 | 0.427 | 0.2724 | Yes |
| 20 | GNPTAB | NA | 2681 | 0.423 | 0.2848 | Yes |
| 21 | AP3D1 | NA | 2825 | 0.416 | 0.2960 | Yes |
| 22 | SNX16 | NA | 2839 | 0.416 | 0.3095 | Yes |
| 23 | MTM1 | NA | 2916 | 0.412 | 0.3217 | Yes |
| 24 | RBSN | NA | 3041 | 0.408 | 0.3329 | Yes |
| 25 | VPS11 | NA | 3042 | 0.408 | 0.3463 | Yes |
| 26 | VPS54 | NA | 3178 | 0.403 | 0.3572 | Yes |
| 27 | SCARB2 | NA | 3276 | 0.399 | 0.3686 | Yes |
| 28 | VPS41 | NA | 3371 | 0.396 | 0.3799 | Yes |
| 29 | VPS35 | NA | 3595 | 0.388 | 0.3886 | Yes |
| 30 | LRP1 | NA | 3758 | 0.382 | 0.3982 | Yes |
| 31 | HOOK2 | NA | 3802 | 0.380 | 0.4100 | Yes |
| 32 | CLEC16A | NA | 3849 | 0.378 | 0.4216 | Yes |
| 33 | VAMP7 | NA | 3864 | 0.378 | 0.4339 | Yes |
| 34 | TMEM106B | NA | 3997 | 0.374 | 0.4438 | Yes |
| 35 | VPS53 | NA | 4159 | 0.369 | 0.4530 | Yes |
| 36 | HGS | NA | 4170 | 0.369 | 0.4650 | Yes |
| 37 | SNX27 | NA | 4474 | 0.360 | 0.4714 | Yes |
| 38 | VPS4A | NA | 4481 | 0.360 | 0.4831 | Yes |
| 39 | AP3B1 | NA | 4493 | 0.360 | 0.4948 | Yes |
| 40 | VPS52 | NA | 4991 | 0.346 | 0.4971 | Yes |
| 41 | EPG5 | NA | 5021 | 0.345 | 0.5080 | Yes |
| 42 | VPS16 | NA | 5144 | 0.342 | 0.5170 | Yes |
| 43 | SORL1 | NA | 5570 | 0.331 | 0.5201 | Yes |
| 44 | GRN | NA | 5685 | 0.328 | 0.5288 | Yes |
| 45 | GGA3 | NA | 5841 | 0.324 | 0.5367 | Yes |
| 46 | CHMP3 | NA | 6062 | 0.319 | 0.5432 | Yes |
| 47 | TRAK2 | NA | 6152 | 0.317 | 0.5520 | Yes |
| 48 | ATG14 | NA | 6374 | 0.312 | 0.5583 | Yes |
| 49 | HOOK3 | NA | 6481 | 0.310 | 0.5666 | Yes |
| 50 | UBXN6 | NA | 6837 | 0.302 | 0.5700 | Yes |
| 51 | SCYL2 | NA | 6851 | 0.302 | 0.5798 | Yes |
| 52 | GCC2 | NA | 6886 | 0.301 | 0.5891 | Yes |
| 53 | PINK1 | NA | 7180 | 0.296 | 0.5935 | Yes |
| 54 | EHD3 | NA | 7201 | 0.295 | 0.6029 | Yes |
| 55 | FAM160A2 | NA | 7241 | 0.294 | 0.6119 | Yes |
| 56 | SPTBN5 | NA | 7384 | 0.291 | 0.6189 | Yes |
| 57 | VPS51 | NA | 7737 | 0.284 | 0.6218 | Yes |
| 58 | M6PR | NA | 8553 | 0.269 | 0.6157 | Yes |
| 59 | RAB12 | NA | 8709 | 0.267 | 0.6217 | Yes |
| 60 | CCDC91 | NA | 8717 | 0.266 | 0.6304 | Yes |
| 61 | USE1 | NA | 8738 | 0.266 | 0.6388 | Yes |
| 62 | PSAP | NA | 8774 | 0.265 | 0.6469 | Yes |
| 63 | BIN1 | NA | 9229 | 0.257 | 0.6470 | Yes |
| 64 | VIPAS39 | NA | 9480 | 0.253 | 0.6508 | Yes |
| 65 | SNAPIN | NA | 9815 | 0.247 | 0.6528 | Yes |
| 66 | ARSB | NA | 10951 | 0.230 | 0.6396 | No |
| 67 | GAK | NA | 11664 | 0.219 | 0.6338 | No |
| 68 | LAMP1 | NA | 12298 | 0.211 | 0.6291 | No |
| 69 | CDX2 | NA | 12825 | 0.203 | 0.6262 | No |
| 70 | MGRN1 | NA | 13296 | 0.196 | 0.6240 | No |
| 71 | HOOK1 | NA | 13374 | 0.195 | 0.6291 | No |
| 72 | VCP | NA | 13405 | 0.195 | 0.6349 | No |
| 73 | AP4M1 | NA | 15375 | 0.169 | 0.6044 | No |
| 74 | DTX3L | NA | 15470 | 0.168 | 0.6082 | No |
| 75 | LRRK2 | NA | 15895 | 0.163 | 0.6058 | No |
| 76 | RAB7B | NA | 16457 | 0.156 | 0.6006 | No |
| 77 | HMGXB4 | NA | 16535 | 0.155 | 0.6043 | No |
| 78 | LAMP2 | NA | 17036 | 0.149 | 0.6001 | No |
| 79 | CLU | NA | 17160 | 0.147 | 0.6027 | No |
| 80 | AKTIP | NA | 17403 | 0.144 | 0.6030 | No |
| 81 | CLN3 | NA | 18819 | 0.126 | 0.5812 | No |
| 82 | TSG101 | NA | 19066 | 0.123 | 0.5807 | No |
| 83 | BECN2 | NA | 20324 | 0.107 | 0.5612 | No |
| 84 | NEDD4 | NA | 20989 | 0.100 | 0.5523 | No |
| 85 | RNF157 | NA | 22726 | 0.080 | 0.5231 | No |
| 86 | CACNG4 | NA | 23131 | 0.076 | 0.5182 | No |
| 87 | GPRASP1 | NA | 23247 | 0.075 | 0.5185 | No |
| 88 | DENND3 | NA | 23519 | 0.072 | 0.5159 | No |
| 89 | NPC1 | NA | 23944 | 0.068 | 0.5104 | No |
| 90 | MVB12A | NA | 24209 | 0.065 | 0.5077 | No |
| 91 | LYST | NA | 29633 | 0.022 | 0.4089 | No |
| 92 | RAB7A | NA | 30138 | 0.020 | 0.4003 | No |
| 93 | CHMP2B | NA | 33749 | 0.003 | 0.3341 | No |
| 94 | HSPA1A | NA | 39308 | -0.003 | 0.2322 | No |
| 95 | HSPA8 | NA | 39913 | -0.011 | 0.2214 | No |
| 96 | PCDHGA3 | NA | 41491 | -0.029 | 0.1935 | No |
| 97 | PCSK9 | NA | 42943 | -0.048 | 0.1684 | No |
| 98 | CHMP5 | NA | 43360 | -0.053 | 0.1625 | No |
| 99 | ADRB2 | NA | 43362 | -0.053 | 0.1643 | No |
| 100 | CACNG7 | NA | 43846 | -0.060 | 0.1574 | No |
| 101 | CACNG2 | NA | 44804 | -0.073 | 0.1422 | No |
| 102 | C9orf72 | NA | 44870 | -0.074 | 0.1435 | No |
| 103 | CACNG5 | NA | 45981 | -0.091 | 0.1261 | No |
| 104 | STX8 | NA | 46000 | -0.091 | 0.1288 | No |
| 105 | RILP | NA | 46105 | -0.093 | 0.1300 | No |
| 106 | LAPTM5 | NA | 46576 | -0.100 | 0.1247 | No |
| 107 | WASH3P | NA | 46600 | -0.101 | 0.1276 | No |
| 108 | ARF1 | NA | 46706 | -0.102 | 0.1290 | No |
| 109 | RHOB | NA | 51638 | -0.206 | 0.0453 | No |
| 110 | CACNG3 | NA | 53420 | -0.269 | 0.0215 | No |