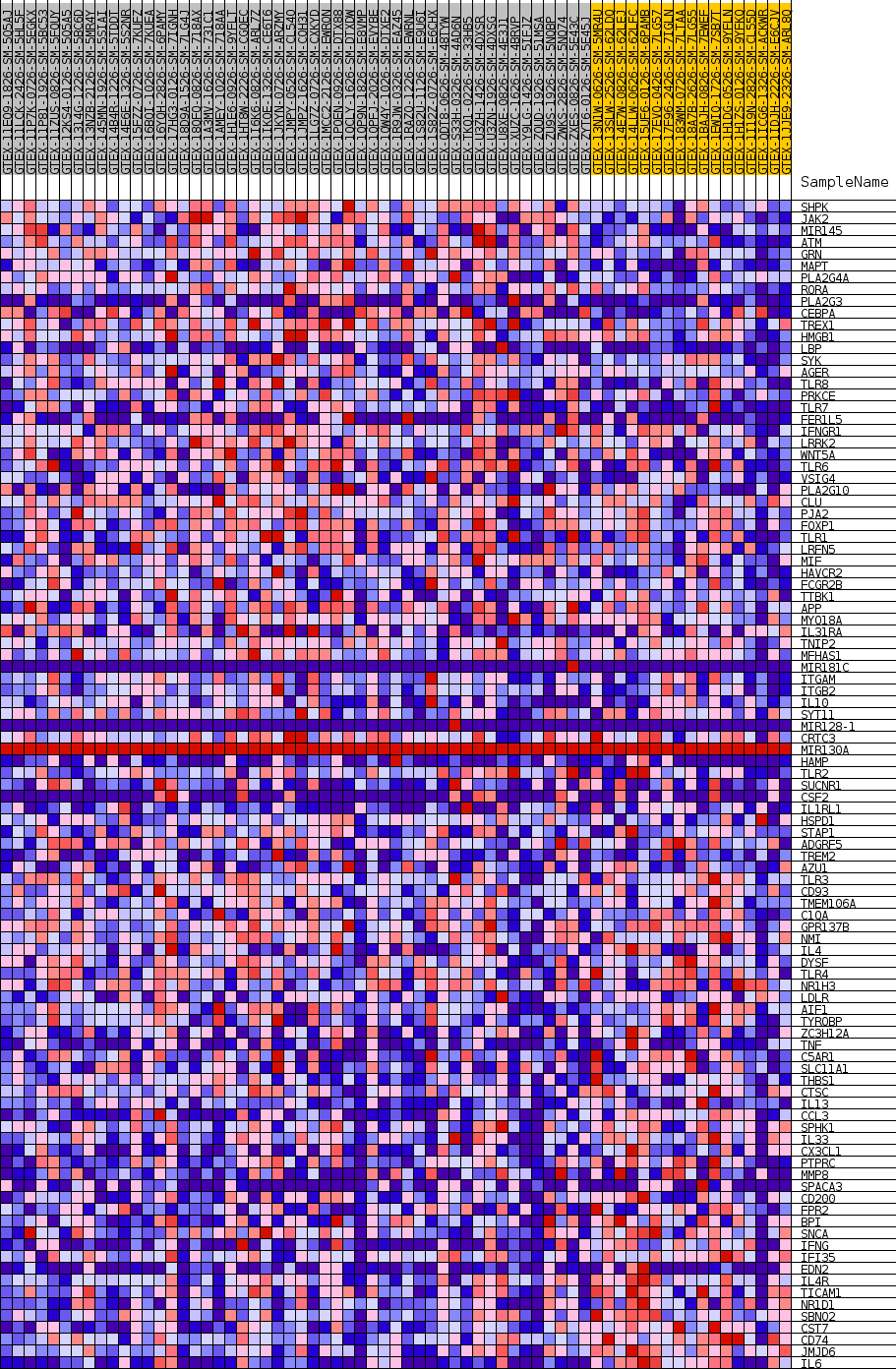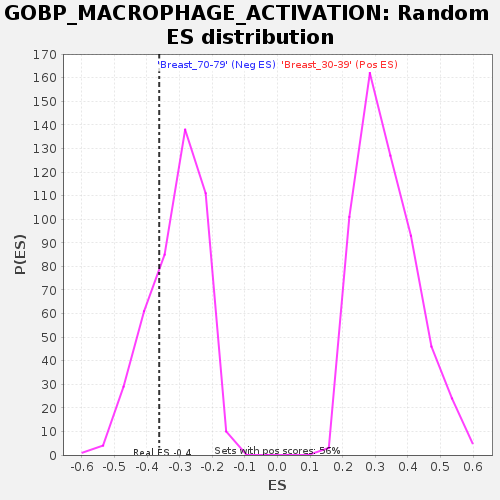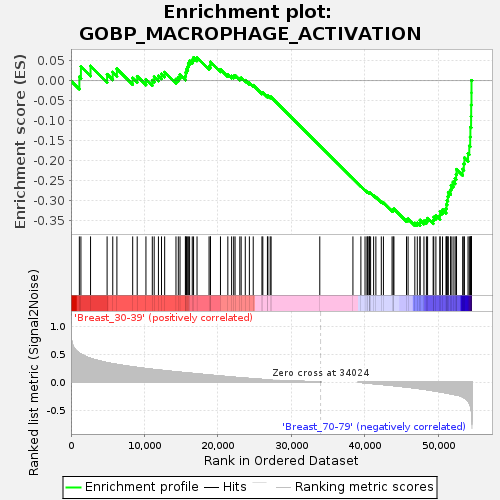
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Breast.Breast.cls #Breast_30-39_versus_Breast_70-79.Breast.cls #Breast_30-39_versus_Breast_70-79_repos |
| Phenotype | Breast.cls#Breast_30-39_versus_Breast_70-79_repos |
| Upregulated in class | Breast_70-79 |
| GeneSet | GOBP_MACROPHAGE_ACTIVATION |
| Enrichment Score (ES) | -0.36255366 |
| Normalized Enrichment Score (NES) | -1.1668245 |
| Nominal p-value | 0.25284737 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.999 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | SHPK | NA | 1137 | 0.517 | 0.0099 | No |
| 2 | JAK2 | NA | 1350 | 0.499 | 0.0358 | No |
| 3 | MIR145 | NA | 2661 | 0.424 | 0.0370 | No |
| 4 | ATM | NA | 4915 | 0.348 | 0.0163 | No |
| 5 | GRN | NA | 5685 | 0.328 | 0.0218 | No |
| 6 | MAPT | NA | 6249 | 0.315 | 0.0302 | No |
| 7 | PLA2G4A | NA | 8399 | 0.272 | 0.0069 | No |
| 8 | RORA | NA | 9013 | 0.261 | 0.0112 | No |
| 9 | PLA2G3 | NA | 10203 | 0.241 | 0.0038 | No |
| 10 | CEBPA | NA | 11069 | 0.228 | 0.0015 | No |
| 11 | TREX1 | NA | 11326 | 0.224 | 0.0102 | No |
| 12 | HMGB1 | NA | 11903 | 0.216 | 0.0124 | No |
| 13 | LBP | NA | 12322 | 0.210 | 0.0173 | No |
| 14 | SYK | NA | 12753 | 0.205 | 0.0216 | No |
| 15 | AGER | NA | 14292 | 0.183 | 0.0043 | No |
| 16 | TLR8 | NA | 14608 | 0.179 | 0.0091 | No |
| 17 | PRKCE | NA | 14842 | 0.176 | 0.0153 | No |
| 18 | TLR7 | NA | 15573 | 0.167 | 0.0118 | No |
| 19 | FER1L5 | NA | 15616 | 0.166 | 0.0210 | No |
| 20 | IFNGR1 | NA | 15729 | 0.165 | 0.0287 | No |
| 21 | LRRK2 | NA | 15895 | 0.163 | 0.0354 | No |
| 22 | WNT5A | NA | 15967 | 0.162 | 0.0437 | No |
| 23 | TLR6 | NA | 16144 | 0.160 | 0.0500 | No |
| 24 | VSIG4 | NA | 16509 | 0.155 | 0.0525 | No |
| 25 | PLA2G10 | NA | 16686 | 0.153 | 0.0584 | No |
| 26 | CLU | NA | 17160 | 0.147 | 0.0585 | No |
| 27 | PJA2 | NA | 18797 | 0.126 | 0.0360 | No |
| 28 | FOXP1 | NA | 18979 | 0.124 | 0.0401 | No |
| 29 | TLR1 | NA | 18985 | 0.124 | 0.0474 | No |
| 30 | LRFN5 | NA | 20351 | 0.107 | 0.0287 | No |
| 31 | MIF | NA | 21353 | 0.095 | 0.0160 | No |
| 32 | HAVCR2 | NA | 21869 | 0.090 | 0.0119 | No |
| 33 | FCGR2B | NA | 22153 | 0.087 | 0.0119 | No |
| 34 | TTBK1 | NA | 22338 | 0.085 | 0.0135 | No |
| 35 | APP | NA | 22986 | 0.077 | 0.0063 | No |
| 36 | MYO18A | NA | 23168 | 0.075 | 0.0074 | No |
| 37 | IL31RA | NA | 23728 | 0.070 | 0.0013 | No |
| 38 | TNIP2 | NA | 24286 | 0.064 | -0.0051 | No |
| 39 | MFHAS1 | NA | 24810 | 0.059 | -0.0111 | No |
| 40 | MIR181C | NA | 25998 | 0.048 | -0.0301 | No |
| 41 | ITGAM | NA | 26116 | 0.047 | -0.0295 | No |
| 42 | ITGB2 | NA | 26744 | 0.041 | -0.0385 | No |
| 43 | IL10 | NA | 26811 | 0.041 | -0.0373 | No |
| 44 | SYT11 | NA | 27119 | 0.038 | -0.0406 | No |
| 45 | MIR128-1 | NA | 27243 | 0.038 | -0.0407 | No |
| 46 | CRTC3 | NA | 33872 | 0.002 | -0.1622 | No |
| 47 | MIR130A | NA | 38385 | 0.000 | -0.2450 | No |
| 48 | HAMP | NA | 39468 | -0.005 | -0.2645 | No |
| 49 | TLR2 | NA | 40032 | -0.012 | -0.2742 | No |
| 50 | SUCNR1 | NA | 40301 | -0.015 | -0.2782 | No |
| 51 | CSF2 | NA | 40356 | -0.016 | -0.2782 | No |
| 52 | IL1RL1 | NA | 40589 | -0.019 | -0.2814 | No |
| 53 | HSPD1 | NA | 40605 | -0.019 | -0.2805 | No |
| 54 | STAP1 | NA | 40610 | -0.019 | -0.2794 | No |
| 55 | ADGRF5 | NA | 40792 | -0.022 | -0.2815 | No |
| 56 | TREM2 | NA | 41202 | -0.026 | -0.2874 | No |
| 57 | AZU1 | NA | 41510 | -0.030 | -0.2913 | No |
| 58 | TLR3 | NA | 42249 | -0.039 | -0.3026 | No |
| 59 | CD93 | NA | 42537 | -0.042 | -0.3053 | No |
| 60 | TMEM106A | NA | 43714 | -0.058 | -0.3234 | No |
| 61 | C1QA | NA | 43873 | -0.060 | -0.3227 | No |
| 62 | GPR137B | NA | 43967 | -0.062 | -0.3208 | No |
| 63 | NMI | NA | 45682 | -0.086 | -0.3471 | No |
| 64 | IL4 | NA | 45895 | -0.090 | -0.3457 | No |
| 65 | DYSF | NA | 46813 | -0.104 | -0.3563 | Yes |
| 66 | TLR4 | NA | 47149 | -0.110 | -0.3559 | Yes |
| 67 | NR1H3 | NA | 47515 | -0.117 | -0.3556 | Yes |
| 68 | LDLR | NA | 47518 | -0.117 | -0.3487 | Yes |
| 69 | AIF1 | NA | 48055 | -0.127 | -0.3510 | Yes |
| 70 | TYROBP | NA | 48389 | -0.132 | -0.3492 | Yes |
| 71 | ZC3H12A | NA | 48550 | -0.136 | -0.3441 | Yes |
| 72 | TNF | NA | 49309 | -0.151 | -0.3490 | Yes |
| 73 | C5AR1 | NA | 49391 | -0.153 | -0.3414 | Yes |
| 74 | SLC11A1 | NA | 49682 | -0.159 | -0.3372 | Yes |
| 75 | THBS1 | NA | 50228 | -0.171 | -0.3370 | Yes |
| 76 | CTSC | NA | 50256 | -0.172 | -0.3273 | Yes |
| 77 | IL13 | NA | 50585 | -0.179 | -0.3227 | Yes |
| 78 | CCL3 | NA | 51058 | -0.191 | -0.3200 | Yes |
| 79 | SPHK1 | NA | 51119 | -0.192 | -0.3096 | Yes |
| 80 | IL33 | NA | 51195 | -0.194 | -0.2995 | Yes |
| 81 | CX3CL1 | NA | 51304 | -0.197 | -0.2897 | Yes |
| 82 | PTPRC | NA | 51361 | -0.198 | -0.2789 | Yes |
| 83 | MMP8 | NA | 51685 | -0.207 | -0.2725 | Yes |
| 84 | SPACA3 | NA | 51750 | -0.209 | -0.2613 | Yes |
| 85 | CD200 | NA | 52020 | -0.216 | -0.2533 | Yes |
| 86 | FPR2 | NA | 52288 | -0.223 | -0.2450 | Yes |
| 87 | BPI | NA | 52435 | -0.227 | -0.2341 | Yes |
| 88 | SNCA | NA | 52490 | -0.228 | -0.2215 | Yes |
| 89 | IFNG | NA | 53327 | -0.263 | -0.2212 | Yes |
| 90 | IFI35 | NA | 53484 | -0.274 | -0.2078 | Yes |
| 91 | EDN2 | NA | 53563 | -0.279 | -0.1926 | Yes |
| 92 | IL4R | NA | 54057 | -0.333 | -0.1818 | Yes |
| 93 | TICAM1 | NA | 54227 | -0.369 | -0.1629 | Yes |
| 94 | NR1D1 | NA | 54350 | -0.410 | -0.1407 | Yes |
| 95 | SBNO2 | NA | 54377 | -0.422 | -0.1160 | Yes |
| 96 | CST7 | NA | 54473 | -0.478 | -0.0893 | Yes |
| 97 | CD74 | NA | 54486 | -0.487 | -0.0605 | Yes |
| 98 | JMJD6 | NA | 54525 | -0.523 | -0.0301 | Yes |
| 99 | IL6 | NA | 54527 | -0.525 | 0.0012 | Yes |