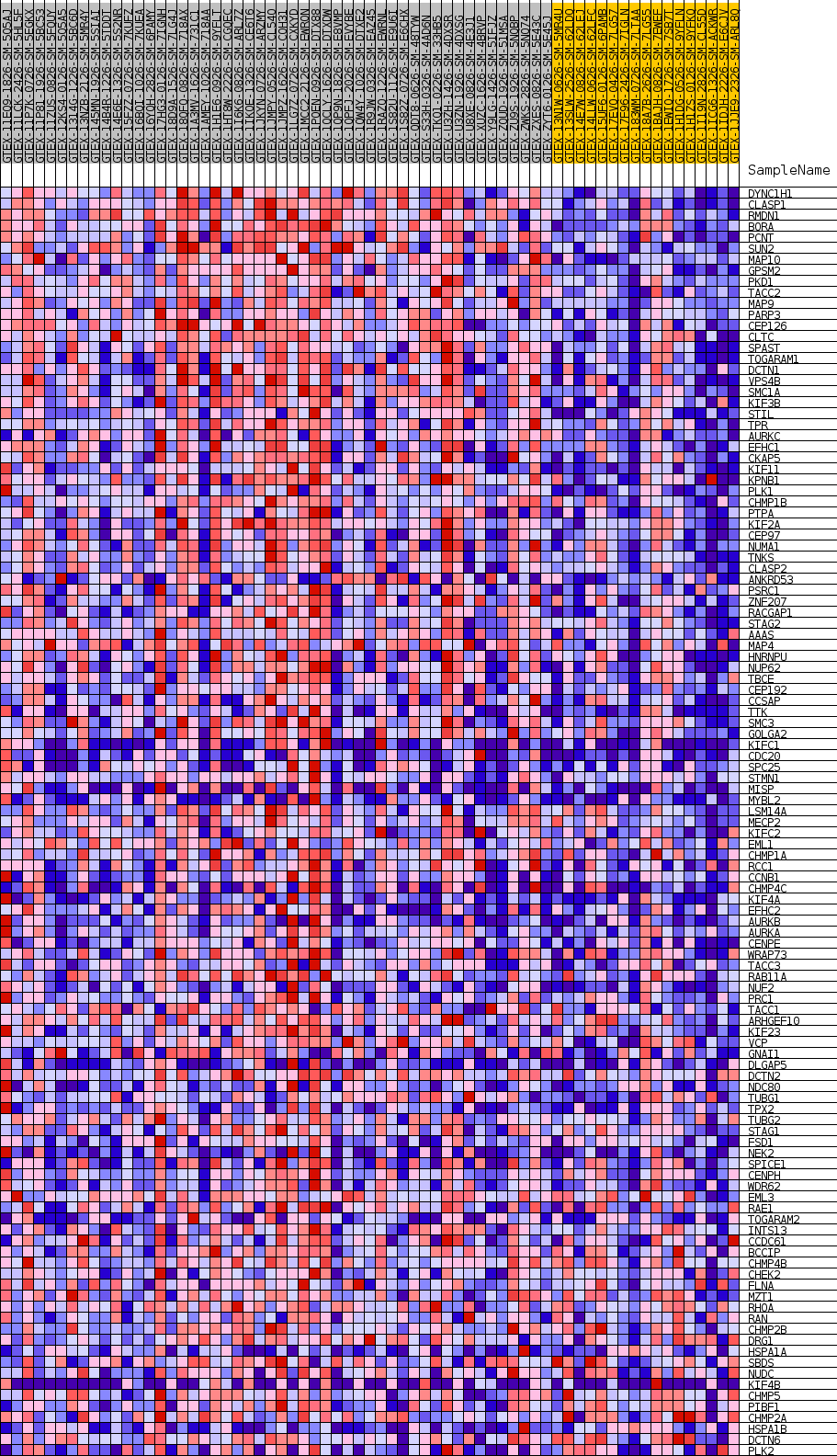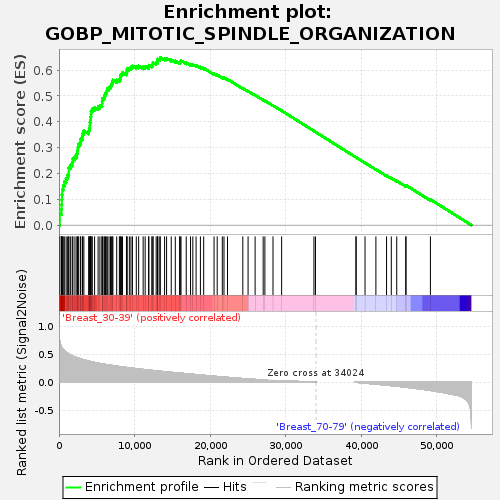
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Breast.Breast.cls #Breast_30-39_versus_Breast_70-79.Breast.cls #Breast_30-39_versus_Breast_70-79_repos |
| Phenotype | Breast.cls#Breast_30-39_versus_Breast_70-79_repos |
| Upregulated in class | Breast_30-39 |
| GeneSet | GOBP_MITOTIC_SPINDLE_ORGANIZATION |
| Enrichment Score (ES) | 0.6487847 |
| Normalized Enrichment Score (NES) | 1.65208 |
| Nominal p-value | 0.0053667263 |
| FDR q-value | 0.2422458 |
| FWER p-Value | 0.795 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | DYNC1H1 | NA | 96 | 0.728 | 0.0213 | Yes |
| 2 | CLASP1 | NA | 98 | 0.728 | 0.0443 | Yes |
| 3 | RMDN1 | NA | 328 | 0.631 | 0.0601 | Yes |
| 4 | BORA | NA | 346 | 0.626 | 0.0796 | Yes |
| 5 | PCNT | NA | 375 | 0.619 | 0.0987 | Yes |
| 6 | SUN2 | NA | 395 | 0.614 | 0.1178 | Yes |
| 7 | MAP10 | NA | 432 | 0.607 | 0.1363 | Yes |
| 8 | GPSM2 | NA | 560 | 0.584 | 0.1525 | Yes |
| 9 | PKD1 | NA | 701 | 0.562 | 0.1677 | Yes |
| 10 | TACC2 | NA | 939 | 0.536 | 0.1804 | Yes |
| 11 | MAP9 | NA | 1108 | 0.520 | 0.1937 | Yes |
| 12 | PARP3 | NA | 1295 | 0.504 | 0.2063 | Yes |
| 13 | CEP126 | NA | 1297 | 0.503 | 0.2222 | Yes |
| 14 | CLTC | NA | 1540 | 0.486 | 0.2331 | Yes |
| 15 | SPAST | NA | 1758 | 0.471 | 0.2441 | Yes |
| 16 | TOGARAM1 | NA | 1841 | 0.466 | 0.2573 | Yes |
| 17 | DCTN1 | NA | 2112 | 0.450 | 0.2666 | Yes |
| 18 | VPS4B | NA | 2362 | 0.438 | 0.2759 | Yes |
| 19 | SMC1A | NA | 2437 | 0.434 | 0.2883 | Yes |
| 20 | KIF3B | NA | 2494 | 0.431 | 0.3009 | Yes |
| 21 | STIL | NA | 2559 | 0.429 | 0.3133 | Yes |
| 22 | TPR | NA | 2817 | 0.417 | 0.3218 | Yes |
| 23 | AURKC | NA | 2880 | 0.413 | 0.3337 | Yes |
| 24 | EFHC1 | NA | 3106 | 0.405 | 0.3424 | Yes |
| 25 | CKAP5 | NA | 3132 | 0.404 | 0.3548 | Yes |
| 26 | KIF11 | NA | 3291 | 0.399 | 0.3645 | Yes |
| 27 | KPNB1 | NA | 3930 | 0.376 | 0.3647 | Yes |
| 28 | PLK1 | NA | 3995 | 0.374 | 0.3754 | Yes |
| 29 | CHMP1B | NA | 4091 | 0.371 | 0.3854 | Yes |
| 30 | PTPA | NA | 4098 | 0.371 | 0.3970 | Yes |
| 31 | KIF2A | NA | 4175 | 0.369 | 0.4073 | Yes |
| 32 | CEP97 | NA | 4185 | 0.369 | 0.4188 | Yes |
| 33 | NUMA1 | NA | 4237 | 0.367 | 0.4295 | Yes |
| 34 | TNKS | NA | 4245 | 0.367 | 0.4410 | Yes |
| 35 | CLASP2 | NA | 4407 | 0.362 | 0.4495 | Yes |
| 36 | ANKRD53 | NA | 4713 | 0.353 | 0.4551 | Yes |
| 37 | PSRC1 | NA | 5175 | 0.341 | 0.4574 | Yes |
| 38 | ZNF207 | NA | 5414 | 0.334 | 0.4636 | Yes |
| 39 | RACGAP1 | NA | 5701 | 0.327 | 0.4688 | Yes |
| 40 | STAG2 | NA | 5711 | 0.327 | 0.4789 | Yes |
| 41 | AAAS | NA | 5728 | 0.327 | 0.4890 | Yes |
| 42 | MAP4 | NA | 5950 | 0.322 | 0.4951 | Yes |
| 43 | HNRNPU | NA | 6042 | 0.320 | 0.5036 | Yes |
| 44 | NUP62 | NA | 6160 | 0.317 | 0.5115 | Yes |
| 45 | TBCE | NA | 6326 | 0.313 | 0.5184 | Yes |
| 46 | CEP192 | NA | 6388 | 0.312 | 0.5271 | Yes |
| 47 | CCSAP | NA | 6620 | 0.307 | 0.5326 | Yes |
| 48 | TTK | NA | 6819 | 0.303 | 0.5385 | Yes |
| 49 | SMC3 | NA | 6935 | 0.300 | 0.5459 | Yes |
| 50 | GOLGA2 | NA | 7054 | 0.298 | 0.5532 | Yes |
| 51 | KIFC1 | NA | 7135 | 0.296 | 0.5611 | Yes |
| 52 | CDC20 | NA | 7637 | 0.286 | 0.5610 | Yes |
| 53 | SPC25 | NA | 7992 | 0.279 | 0.5633 | Yes |
| 54 | STMN1 | NA | 8103 | 0.277 | 0.5701 | Yes |
| 55 | MISP | NA | 8119 | 0.277 | 0.5786 | Yes |
| 56 | MYBL2 | NA | 8265 | 0.274 | 0.5846 | Yes |
| 57 | LSM14A | NA | 8409 | 0.272 | 0.5906 | Yes |
| 58 | MECP2 | NA | 8932 | 0.262 | 0.5893 | Yes |
| 59 | KIFC2 | NA | 8990 | 0.262 | 0.5965 | Yes |
| 60 | EML1 | NA | 9004 | 0.261 | 0.6046 | Yes |
| 61 | CHMP1A | NA | 9313 | 0.256 | 0.6070 | Yes |
| 62 | RCC1 | NA | 9525 | 0.252 | 0.6111 | Yes |
| 63 | CCNB1 | NA | 9721 | 0.249 | 0.6154 | Yes |
| 64 | CHMP4C | NA | 10232 | 0.241 | 0.6137 | Yes |
| 65 | KIF4A | NA | 10542 | 0.236 | 0.6155 | Yes |
| 66 | EFHC2 | NA | 11139 | 0.227 | 0.6117 | Yes |
| 67 | AURKB | NA | 11405 | 0.223 | 0.6139 | Yes |
| 68 | AURKA | NA | 11860 | 0.216 | 0.6124 | Yes |
| 69 | CENPE | NA | 11898 | 0.216 | 0.6186 | Yes |
| 70 | WRAP73 | NA | 12281 | 0.211 | 0.6183 | Yes |
| 71 | TACC3 | NA | 12427 | 0.209 | 0.6222 | Yes |
| 72 | RAB11A | NA | 12447 | 0.209 | 0.6285 | Yes |
| 73 | NUF2 | NA | 12861 | 0.203 | 0.6273 | Yes |
| 74 | PRC1 | NA | 13024 | 0.200 | 0.6307 | Yes |
| 75 | TACC1 | NA | 13069 | 0.200 | 0.6362 | Yes |
| 76 | ARHGEF10 | NA | 13079 | 0.200 | 0.6424 | Yes |
| 77 | KIF23 | NA | 13349 | 0.196 | 0.6436 | Yes |
| 78 | VCP | NA | 13405 | 0.195 | 0.6488 | Yes |
| 79 | GNAI1 | NA | 13977 | 0.187 | 0.6442 | No |
| 80 | DLGAP5 | NA | 14241 | 0.183 | 0.6452 | No |
| 81 | DCTN2 | NA | 14857 | 0.176 | 0.6395 | No |
| 82 | NDC80 | NA | 15394 | 0.169 | 0.6350 | No |
| 83 | TUBG1 | NA | 15962 | 0.162 | 0.6297 | No |
| 84 | TPX2 | NA | 16050 | 0.161 | 0.6332 | No |
| 85 | TUBG2 | NA | 16150 | 0.159 | 0.6364 | No |
| 86 | STAG1 | NA | 16843 | 0.151 | 0.6285 | No |
| 87 | FSD1 | NA | 17413 | 0.144 | 0.6226 | No |
| 88 | NEK2 | NA | 17715 | 0.140 | 0.6215 | No |
| 89 | SPICE1 | NA | 18146 | 0.135 | 0.6179 | No |
| 90 | CENPH | NA | 18718 | 0.127 | 0.6115 | No |
| 91 | WDR62 | NA | 19154 | 0.122 | 0.6073 | No |
| 92 | EML3 | NA | 20527 | 0.105 | 0.5855 | No |
| 93 | RAE1 | NA | 20943 | 0.100 | 0.5810 | No |
| 94 | TOGARAM2 | NA | 21619 | 0.092 | 0.5716 | No |
| 95 | INTS13 | NA | 21840 | 0.090 | 0.5704 | No |
| 96 | CCDC61 | NA | 22303 | 0.085 | 0.5646 | No |
| 97 | BCCIP | NA | 24333 | 0.064 | 0.5294 | No |
| 98 | CHMP4B | NA | 25030 | 0.057 | 0.5184 | No |
| 99 | CHEK2 | NA | 25961 | 0.048 | 0.5028 | No |
| 100 | FLNA | NA | 27028 | 0.039 | 0.4845 | No |
| 101 | MZT1 | NA | 27255 | 0.037 | 0.4815 | No |
| 102 | RHOA | NA | 28328 | 0.030 | 0.4628 | No |
| 103 | RAN | NA | 29471 | 0.023 | 0.4426 | No |
| 104 | CHMP2B | NA | 33749 | 0.003 | 0.3642 | No |
| 105 | DRG1 | NA | 33950 | 0.001 | 0.3605 | No |
| 106 | HSPA1A | NA | 39308 | -0.003 | 0.2623 | No |
| 107 | SBDS | NA | 39336 | -0.003 | 0.2619 | No |
| 108 | NUDC | NA | 40511 | -0.018 | 0.2409 | No |
| 109 | KIF4B | NA | 41950 | -0.035 | 0.2156 | No |
| 110 | CHMP5 | NA | 43360 | -0.053 | 0.1914 | No |
| 111 | PIBF1 | NA | 43989 | -0.062 | 0.1819 | No |
| 112 | CHMP2A | NA | 44709 | -0.072 | 0.1709 | No |
| 113 | HSPA1B | NA | 45902 | -0.090 | 0.1519 | No |
| 114 | DCTN6 | NA | 45939 | -0.090 | 0.1541 | No |
| 115 | PLK2 | NA | 49168 | -0.148 | 0.0995 | No |