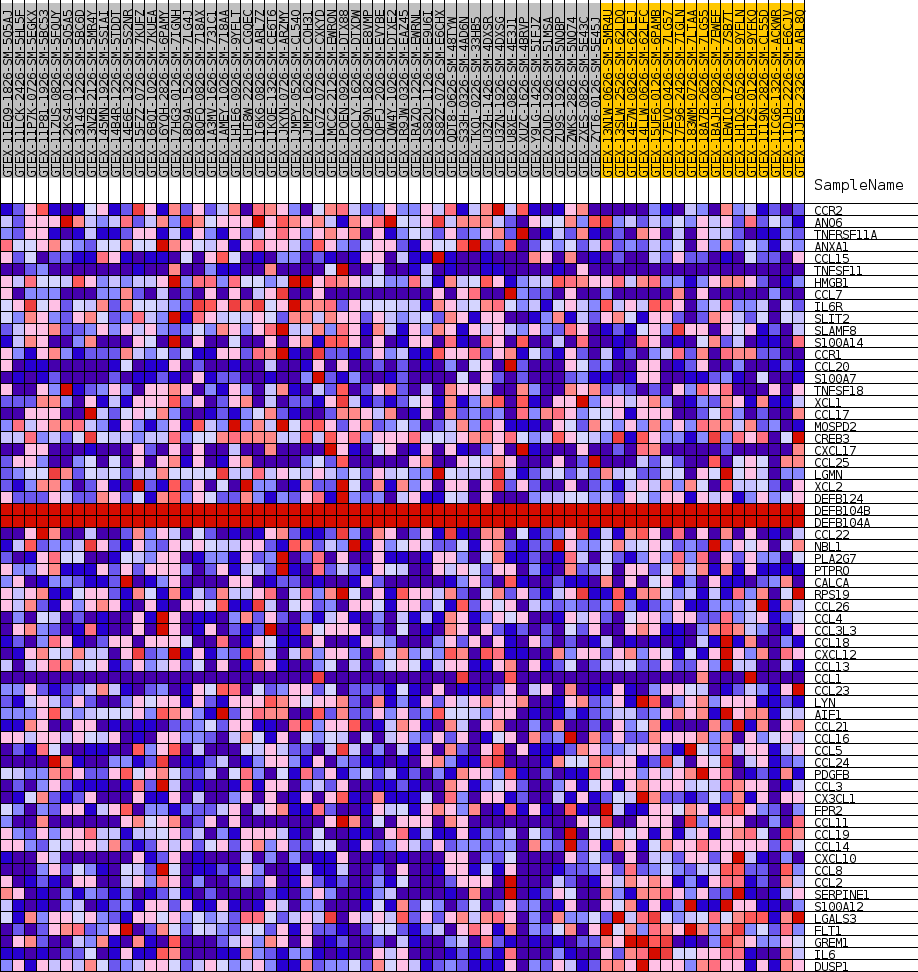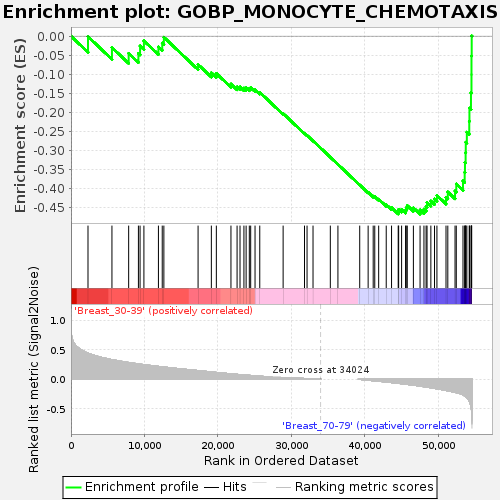
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Breast.Breast.cls #Breast_30-39_versus_Breast_70-79.Breast.cls #Breast_30-39_versus_Breast_70-79_repos |
| Phenotype | Breast.cls#Breast_30-39_versus_Breast_70-79_repos |
| Upregulated in class | Breast_70-79 |
| GeneSet | GOBP_MONOCYTE_CHEMOTAXIS |
| Enrichment Score (ES) | -0.4686351 |
| Normalized Enrichment Score (NES) | -1.3944415 |
| Nominal p-value | 0.085158154 |
| FDR q-value | 0.9160426 |
| FWER p-Value | 0.987 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | CCR2 | NA | 2314 | 0.440 | -0.0012 | No |
| 2 | ANO6 | NA | 5574 | 0.330 | -0.0300 | No |
| 3 | TNFRSF11A | NA | 7851 | 0.282 | -0.0453 | No |
| 4 | ANXA1 | NA | 9178 | 0.258 | -0.0454 | No |
| 5 | CCL15 | NA | 9403 | 0.254 | -0.0257 | No |
| 6 | TNFSF11 | NA | 9926 | 0.246 | -0.0122 | No |
| 7 | HMGB1 | NA | 11903 | 0.216 | -0.0282 | No |
| 8 | CCL7 | NA | 12422 | 0.209 | -0.0181 | No |
| 9 | IL6R | NA | 12644 | 0.206 | -0.0028 | No |
| 10 | SLIT2 | NA | 17302 | 0.145 | -0.0746 | No |
| 11 | SLAMF8 | NA | 19109 | 0.122 | -0.0963 | No |
| 12 | S100A14 | NA | 19789 | 0.114 | -0.0980 | No |
| 13 | CCR1 | NA | 21778 | 0.090 | -0.1260 | No |
| 14 | CCL20 | NA | 22612 | 0.081 | -0.1337 | No |
| 15 | S100A7 | NA | 23012 | 0.077 | -0.1337 | No |
| 16 | TNFSF18 | NA | 23544 | 0.071 | -0.1368 | No |
| 17 | XCL1 | NA | 23843 | 0.068 | -0.1358 | No |
| 18 | CCL17 | NA | 24278 | 0.064 | -0.1378 | No |
| 19 | MOSPD2 | NA | 24464 | 0.063 | -0.1353 | No |
| 20 | CREB3 | NA | 25057 | 0.057 | -0.1408 | No |
| 21 | CXCL17 | NA | 25701 | 0.050 | -0.1479 | No |
| 22 | CCL25 | NA | 28881 | 0.027 | -0.2037 | No |
| 23 | LGMN | NA | 31792 | 0.012 | -0.2559 | No |
| 24 | XCL2 | NA | 32139 | 0.010 | -0.2613 | No |
| 25 | DEFB124 | NA | 32952 | 0.007 | -0.2756 | No |
| 26 | DEFB104B | NA | 35307 | 0.000 | -0.3187 | No |
| 27 | DEFB104A | NA | 36342 | 0.000 | -0.3377 | No |
| 28 | CCL22 | NA | 39310 | -0.003 | -0.3919 | No |
| 29 | NBL1 | NA | 40456 | -0.017 | -0.4112 | No |
| 30 | PLA2G7 | NA | 41144 | -0.025 | -0.4214 | No |
| 31 | PTPRO | NA | 41358 | -0.028 | -0.4227 | No |
| 32 | CALCA | NA | 41890 | -0.034 | -0.4293 | No |
| 33 | RPS19 | NA | 42919 | -0.048 | -0.4437 | No |
| 34 | CCL26 | NA | 43644 | -0.057 | -0.4516 | No |
| 35 | CCL4 | NA | 44550 | -0.070 | -0.4617 | Yes |
| 36 | CCL3L3 | NA | 44612 | -0.071 | -0.4561 | Yes |
| 37 | CCL18 | NA | 45013 | -0.077 | -0.4563 | Yes |
| 38 | CXCL12 | NA | 45552 | -0.084 | -0.4583 | Yes |
| 39 | CCL13 | NA | 45642 | -0.086 | -0.4518 | Yes |
| 40 | CCL1 | NA | 45784 | -0.088 | -0.4462 | Yes |
| 41 | CCL23 | NA | 46617 | -0.101 | -0.4520 | Yes |
| 42 | LYN | NA | 47525 | -0.117 | -0.4577 | Yes |
| 43 | AIF1 | NA | 48055 | -0.127 | -0.4555 | Yes |
| 44 | CCL21 | NA | 48347 | -0.132 | -0.4485 | Yes |
| 45 | CCL16 | NA | 48475 | -0.135 | -0.4382 | Yes |
| 46 | CCL5 | NA | 48995 | -0.145 | -0.4342 | Yes |
| 47 | CCL24 | NA | 49494 | -0.155 | -0.4288 | Yes |
| 48 | PDGFB | NA | 49822 | -0.162 | -0.4196 | Yes |
| 49 | CCL3 | NA | 51058 | -0.191 | -0.4244 | Yes |
| 50 | CX3CL1 | NA | 51304 | -0.197 | -0.4104 | Yes |
| 51 | FPR2 | NA | 52288 | -0.223 | -0.4075 | Yes |
| 52 | CCL11 | NA | 52457 | -0.227 | -0.3893 | Yes |
| 53 | CCL19 | NA | 53370 | -0.266 | -0.3811 | Yes |
| 54 | CCL14 | NA | 53600 | -0.283 | -0.3587 | Yes |
| 55 | CXCL10 | NA | 53640 | -0.286 | -0.3326 | Yes |
| 56 | CCL8 | NA | 53728 | -0.293 | -0.3067 | Yes |
| 57 | CCL2 | NA | 53749 | -0.296 | -0.2793 | Yes |
| 58 | SERPINE1 | NA | 53895 | -0.311 | -0.2528 | Yes |
| 59 | S100A12 | NA | 54225 | -0.369 | -0.2243 | Yes |
| 60 | LGALS3 | NA | 54248 | -0.375 | -0.1895 | Yes |
| 61 | FLT1 | NA | 54457 | -0.470 | -0.1492 | Yes |
| 62 | GREM1 | NA | 54526 | -0.525 | -0.1012 | Yes |
| 63 | IL6 | NA | 54527 | -0.525 | -0.0520 | Yes |
| 64 | DUSP1 | NA | 54556 | -0.567 | 0.0006 | Yes |