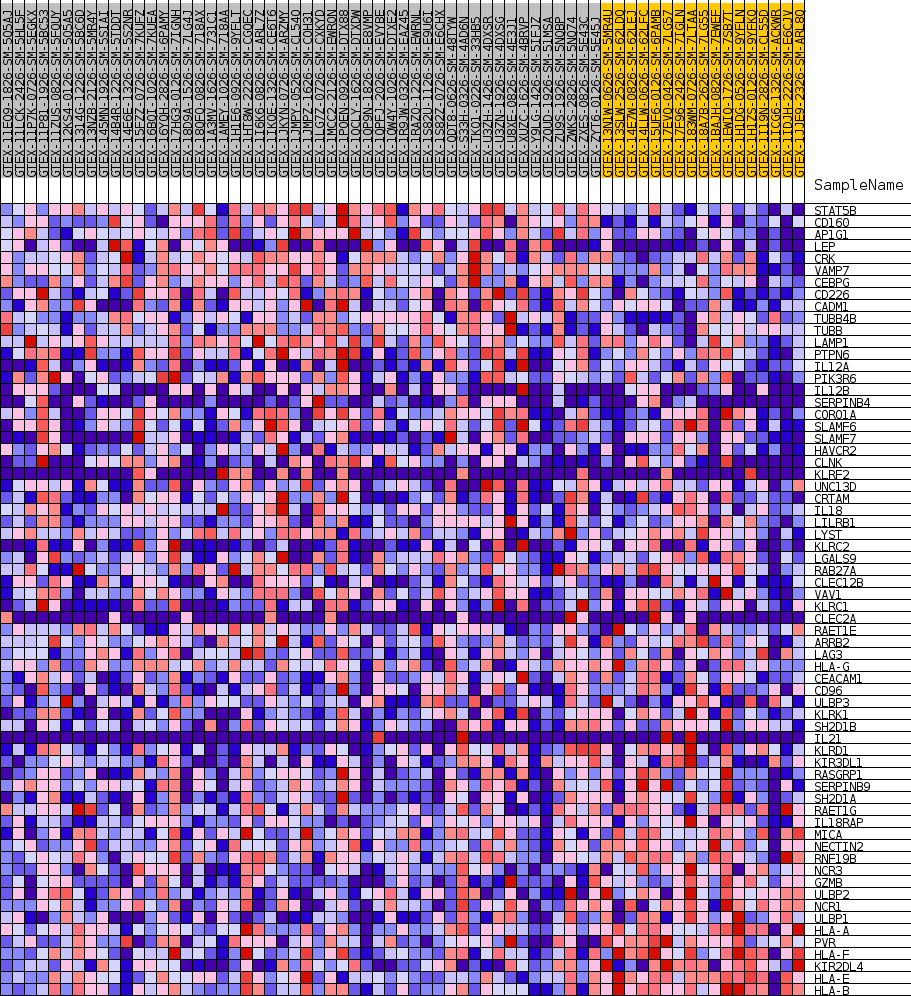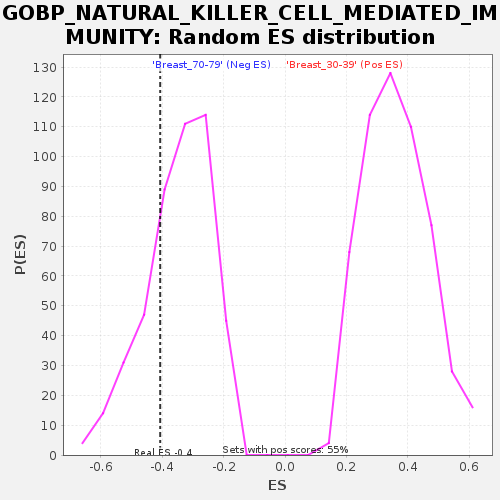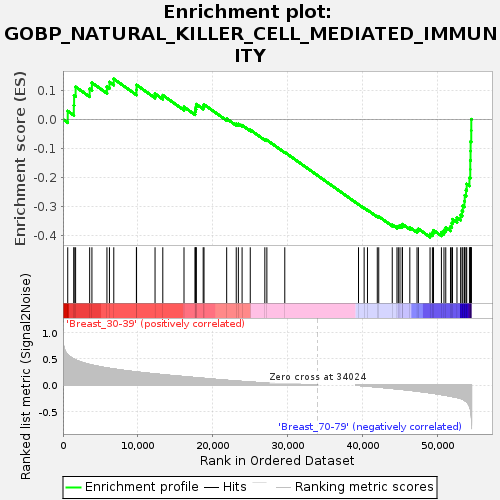
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Breast.Breast.cls #Breast_30-39_versus_Breast_70-79.Breast.cls #Breast_30-39_versus_Breast_70-79_repos |
| Phenotype | Breast.cls#Breast_30-39_versus_Breast_70-79_repos |
| Upregulated in class | Breast_70-79 |
| GeneSet | GOBP_NATURAL_KILLER_CELL_MEDIATED_IMMUNITY |
| Enrichment Score (ES) | -0.40677667 |
| Normalized Enrichment Score (NES) | -1.1691608 |
| Nominal p-value | 0.26153848 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.999 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | STAT5B | NA | 632 | 0.572 | 0.0288 | No |
| 2 | CD160 | NA | 1447 | 0.492 | 0.0487 | No |
| 3 | AP1G1 | NA | 1483 | 0.489 | 0.0826 | No |
| 4 | LEP | NA | 1684 | 0.477 | 0.1127 | No |
| 5 | CRK | NA | 3562 | 0.389 | 0.1057 | No |
| 6 | VAMP7 | NA | 3864 | 0.378 | 0.1269 | No |
| 7 | CEBPG | NA | 5871 | 0.323 | 0.1130 | No |
| 8 | CD226 | NA | 6206 | 0.316 | 0.1292 | No |
| 9 | CADM1 | NA | 6785 | 0.303 | 0.1400 | No |
| 10 | TUBB4B | NA | 9807 | 0.248 | 0.1021 | No |
| 11 | TUBB | NA | 9829 | 0.247 | 0.1192 | No |
| 12 | LAMP1 | NA | 12298 | 0.211 | 0.0889 | No |
| 13 | PTPN6 | NA | 13334 | 0.196 | 0.0837 | No |
| 14 | IL12A | NA | 16167 | 0.159 | 0.0430 | No |
| 15 | PIK3R6 | NA | 17640 | 0.141 | 0.0260 | No |
| 16 | IL12B | NA | 17687 | 0.141 | 0.0351 | No |
| 17 | SERPINB4 | NA | 17772 | 0.140 | 0.0435 | No |
| 18 | CORO1A | NA | 17805 | 0.139 | 0.0527 | No |
| 19 | SLAMF6 | NA | 18730 | 0.127 | 0.0447 | No |
| 20 | SLAMF7 | NA | 18841 | 0.126 | 0.0516 | No |
| 21 | HAVCR2 | NA | 21869 | 0.090 | 0.0024 | No |
| 22 | CLNK | NA | 23147 | 0.076 | -0.0156 | No |
| 23 | KLRF2 | NA | 23430 | 0.073 | -0.0157 | No |
| 24 | UNC13D | NA | 23928 | 0.068 | -0.0200 | No |
| 25 | CRTAM | NA | 25021 | 0.057 | -0.0360 | No |
| 26 | IL18 | NA | 26968 | 0.040 | -0.0689 | No |
| 27 | LILRB1 | NA | 27240 | 0.038 | -0.0712 | No |
| 28 | LYST | NA | 29633 | 0.022 | -0.1135 | No |
| 29 | KLRC2 | NA | 39484 | -0.005 | -0.2938 | No |
| 30 | LGALS9 | NA | 40236 | -0.015 | -0.3065 | No |
| 31 | RAB27A | NA | 40682 | -0.020 | -0.3133 | No |
| 32 | CLEC12B | NA | 41992 | -0.035 | -0.3348 | No |
| 33 | VAV1 | NA | 42188 | -0.038 | -0.3357 | No |
| 34 | KLRC1 | NA | 43992 | -0.062 | -0.3644 | No |
| 35 | CLEC2A | NA | 44623 | -0.071 | -0.3709 | No |
| 36 | RAET1E | NA | 44805 | -0.073 | -0.3691 | No |
| 37 | ARRB2 | NA | 45025 | -0.077 | -0.3676 | No |
| 38 | LAG3 | NA | 45326 | -0.081 | -0.3674 | No |
| 39 | HLA-G | NA | 45353 | -0.081 | -0.3622 | No |
| 40 | CEACAM1 | NA | 46340 | -0.097 | -0.3734 | No |
| 41 | CD96 | NA | 47302 | -0.112 | -0.3831 | No |
| 42 | ULBP3 | NA | 47487 | -0.116 | -0.3783 | No |
| 43 | KLRK1 | NA | 49043 | -0.146 | -0.3965 | Yes |
| 44 | SH2D1B | NA | 49358 | -0.152 | -0.3915 | Yes |
| 45 | IL21 | NA | 49506 | -0.155 | -0.3832 | Yes |
| 46 | KLRD1 | NA | 50561 | -0.178 | -0.3899 | Yes |
| 47 | KIR3DL1 | NA | 50903 | -0.187 | -0.3830 | Yes |
| 48 | RASGRP1 | NA | 51143 | -0.193 | -0.3737 | Yes |
| 49 | SERPINB9 | NA | 51773 | -0.209 | -0.3705 | Yes |
| 50 | SH2D1A | NA | 51946 | -0.213 | -0.3586 | Yes |
| 51 | RAET1G | NA | 52044 | -0.216 | -0.3451 | Yes |
| 52 | IL18RAP | NA | 52639 | -0.233 | -0.3395 | Yes |
| 53 | MICA | NA | 53128 | -0.251 | -0.3307 | Yes |
| 54 | NECTIN2 | NA | 53322 | -0.262 | -0.3157 | Yes |
| 55 | RNF19B | NA | 53427 | -0.270 | -0.2985 | Yes |
| 56 | NCR3 | NA | 53635 | -0.286 | -0.2821 | Yes |
| 57 | GZMB | NA | 53679 | -0.290 | -0.2624 | Yes |
| 58 | ULBP2 | NA | 53851 | -0.306 | -0.2439 | Yes |
| 59 | NCR1 | NA | 53943 | -0.317 | -0.2232 | Yes |
| 60 | ULBP1 | NA | 54305 | -0.394 | -0.2019 | Yes |
| 61 | HLA-A | NA | 54414 | -0.439 | -0.1729 | Yes |
| 62 | PVR | NA | 54417 | -0.441 | -0.1417 | Yes |
| 63 | HLA-F | NA | 54455 | -0.467 | -0.1094 | Yes |
| 64 | KIR2DL4 | NA | 54465 | -0.472 | -0.0762 | Yes |
| 65 | HLA-E | NA | 54547 | -0.553 | -0.0386 | Yes |
| 66 | HLA-B | NA | 54552 | -0.557 | 0.0007 | Yes |