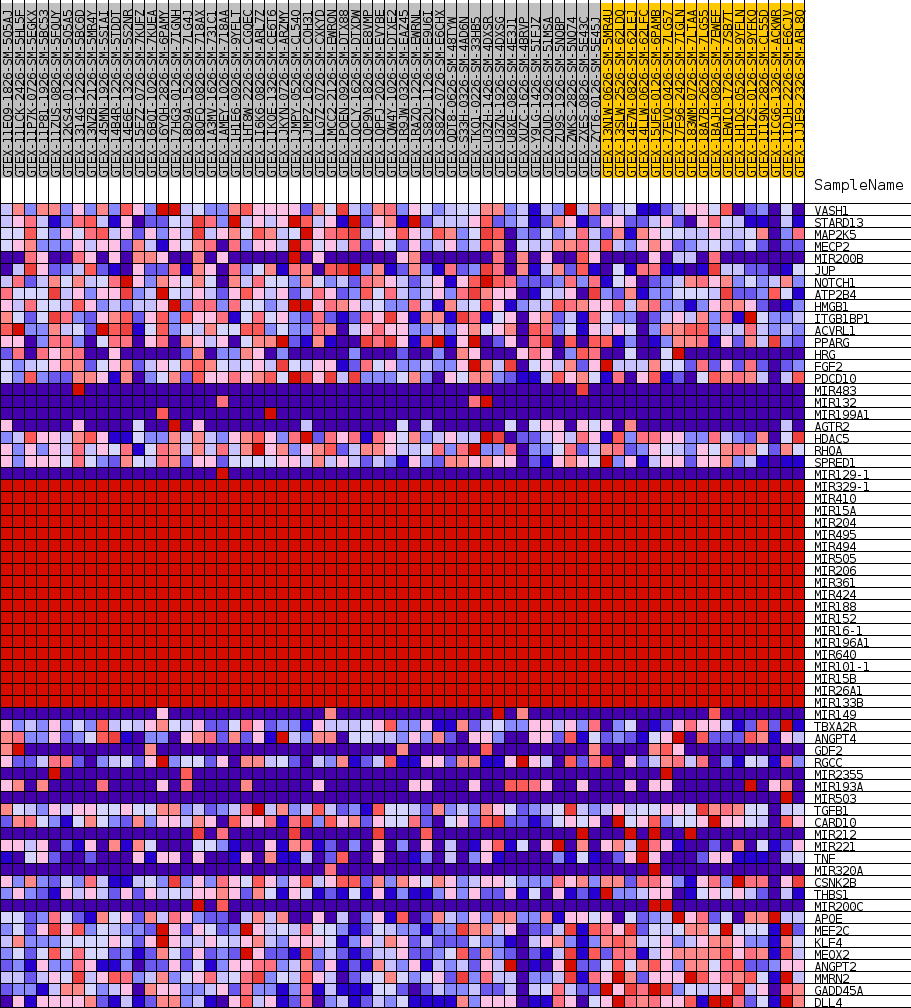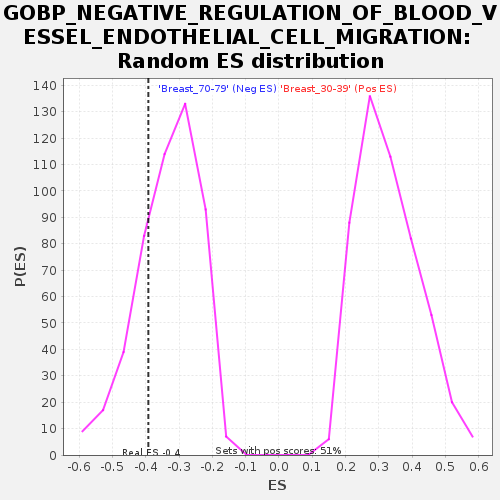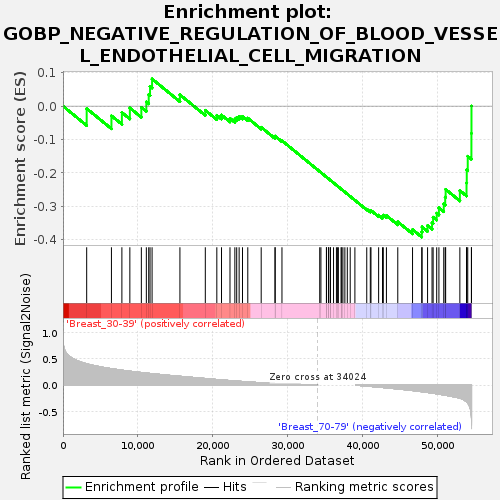
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Breast.Breast.cls #Breast_30-39_versus_Breast_70-79.Breast.cls #Breast_30-39_versus_Breast_70-79_repos |
| Phenotype | Breast.cls#Breast_30-39_versus_Breast_70-79_repos |
| Upregulated in class | Breast_70-79 |
| GeneSet | GOBP_NEGATIVE_REGULATION_OF_BLOOD_VESSEL_ENDOTHELIAL_CELL_MIGRATION |
| Enrichment Score (ES) | -0.39218447 |
| Normalized Enrichment Score (NES) | -1.1789017 |
| Nominal p-value | 0.24040404 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.999 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | VASH1 | NA | 3164 | 0.403 | -0.0072 | No |
| 2 | STARD13 | NA | 6471 | 0.310 | -0.0289 | No |
| 3 | MAP2K5 | NA | 7868 | 0.282 | -0.0190 | No |
| 4 | MECP2 | NA | 8932 | 0.262 | -0.0054 | No |
| 5 | MIR200B | NA | 10468 | 0.237 | -0.0037 | No |
| 6 | JUP | NA | 11135 | 0.227 | 0.0126 | No |
| 7 | NOTCH1 | NA | 11453 | 0.222 | 0.0348 | No |
| 8 | ATP2B4 | NA | 11629 | 0.220 | 0.0593 | No |
| 9 | HMGB1 | NA | 11903 | 0.216 | 0.0815 | No |
| 10 | ITGB1BP1 | NA | 15622 | 0.166 | 0.0342 | No |
| 11 | ACVRL1 | NA | 19014 | 0.124 | -0.0124 | No |
| 12 | PPARG | NA | 20553 | 0.105 | -0.0274 | No |
| 13 | HRG | NA | 21179 | 0.098 | -0.0266 | No |
| 14 | FGF2 | NA | 22312 | 0.085 | -0.0367 | No |
| 15 | PDCD10 | NA | 22962 | 0.078 | -0.0388 | No |
| 16 | MIR483 | NA | 23209 | 0.075 | -0.0339 | No |
| 17 | MIR132 | NA | 23534 | 0.072 | -0.0308 | No |
| 18 | MIR199A1 | NA | 23983 | 0.067 | -0.0306 | No |
| 19 | AGTR2 | NA | 24690 | 0.060 | -0.0359 | No |
| 20 | HDAC5 | NA | 26481 | 0.043 | -0.0633 | No |
| 21 | RHOA | NA | 28328 | 0.030 | -0.0933 | No |
| 22 | SPRED1 | NA | 28347 | 0.030 | -0.0898 | No |
| 23 | MIR129-1 | NA | 29256 | 0.025 | -0.1034 | No |
| 24 | MIR329-1 | NA | 34302 | 0.000 | -0.1959 | No |
| 25 | MIR410 | NA | 34461 | 0.000 | -0.1988 | No |
| 26 | MIR15A | NA | 35194 | 0.000 | -0.2123 | No |
| 27 | MIR204 | NA | 35214 | 0.000 | -0.2126 | No |
| 28 | MIR495 | NA | 35480 | 0.000 | -0.2175 | No |
| 29 | MIR494 | NA | 35493 | 0.000 | -0.2177 | No |
| 30 | MIR505 | NA | 35711 | 0.000 | -0.2217 | No |
| 31 | MIR206 | NA | 35744 | 0.000 | -0.2222 | No |
| 32 | MIR361 | NA | 36147 | 0.000 | -0.2296 | No |
| 33 | MIR424 | NA | 36533 | 0.000 | -0.2367 | No |
| 34 | MIR188 | NA | 36665 | 0.000 | -0.2391 | No |
| 35 | MIR152 | NA | 36795 | 0.000 | -0.2414 | No |
| 36 | MIR16-1 | NA | 37152 | 0.000 | -0.2480 | No |
| 37 | MIR196A1 | NA | 37238 | 0.000 | -0.2495 | No |
| 38 | MIR640 | NA | 37399 | 0.000 | -0.2525 | No |
| 39 | MIR101-1 | NA | 37661 | 0.000 | -0.2573 | No |
| 40 | MIR15B | NA | 37987 | 0.000 | -0.2632 | No |
| 41 | MIR26A1 | NA | 38376 | 0.000 | -0.2703 | No |
| 42 | MIR133B | NA | 39001 | 0.000 | -0.2818 | No |
| 43 | MIR149 | NA | 40585 | -0.019 | -0.3085 | No |
| 44 | TBXA2R | NA | 41059 | -0.025 | -0.3140 | No |
| 45 | ANGPT4 | NA | 41150 | -0.026 | -0.3125 | No |
| 46 | GDF2 | NA | 42172 | -0.038 | -0.3265 | No |
| 47 | RGCC | NA | 42690 | -0.044 | -0.3304 | No |
| 48 | MIR2355 | NA | 42796 | -0.046 | -0.3265 | No |
| 49 | MIR193A | NA | 43213 | -0.051 | -0.3277 | No |
| 50 | MIR503 | NA | 44731 | -0.073 | -0.3464 | No |
| 51 | TGFB1 | NA | 46703 | -0.102 | -0.3696 | No |
| 52 | CARD10 | NA | 47934 | -0.124 | -0.3765 | Yes |
| 53 | MIR212 | NA | 47978 | -0.125 | -0.3616 | Yes |
| 54 | MIR221 | NA | 48712 | -0.139 | -0.3575 | Yes |
| 55 | TNF | NA | 49309 | -0.151 | -0.3494 | Yes |
| 56 | MIR320A | NA | 49453 | -0.154 | -0.3326 | Yes |
| 57 | CSNK2B | NA | 49931 | -0.165 | -0.3206 | Yes |
| 58 | THBS1 | NA | 50228 | -0.171 | -0.3044 | Yes |
| 59 | MIR200C | NA | 50873 | -0.186 | -0.2928 | Yes |
| 60 | APOE | NA | 51066 | -0.191 | -0.2723 | Yes |
| 61 | MEF2C | NA | 51127 | -0.192 | -0.2492 | Yes |
| 62 | KLF4 | NA | 53026 | -0.247 | -0.2529 | Yes |
| 63 | MEOX2 | NA | 53916 | -0.313 | -0.2297 | Yes |
| 64 | ANGPT2 | NA | 53948 | -0.318 | -0.1903 | Yes |
| 65 | MMRN2 | NA | 54079 | -0.338 | -0.1501 | Yes |
| 66 | GADD45A | NA | 54569 | -0.614 | -0.0817 | Yes |
| 67 | DLL4 | NA | 54578 | -0.652 | 0.0002 | Yes |