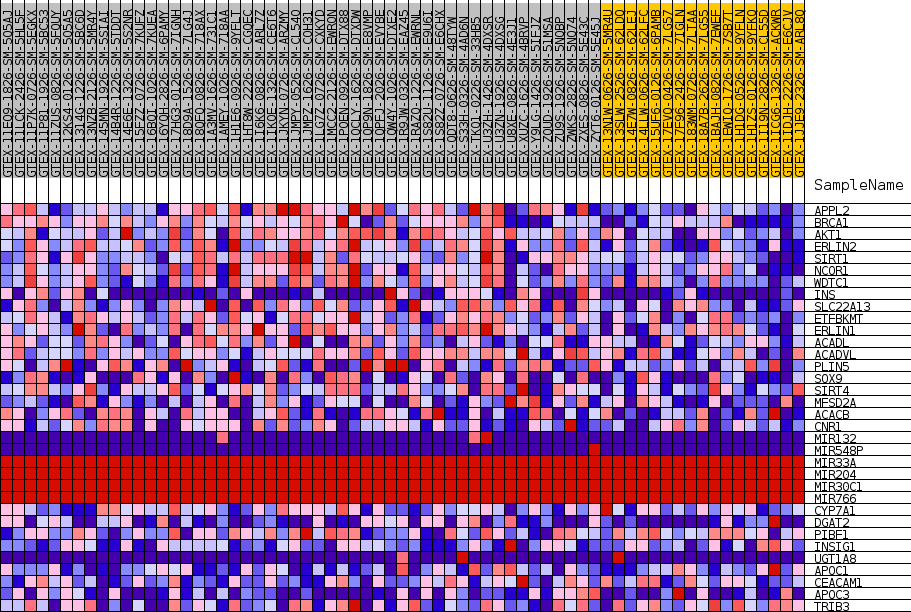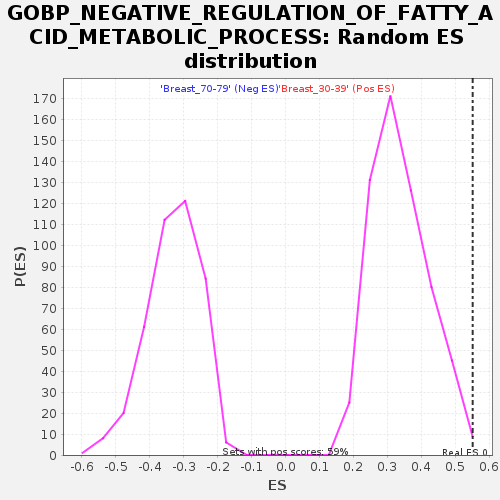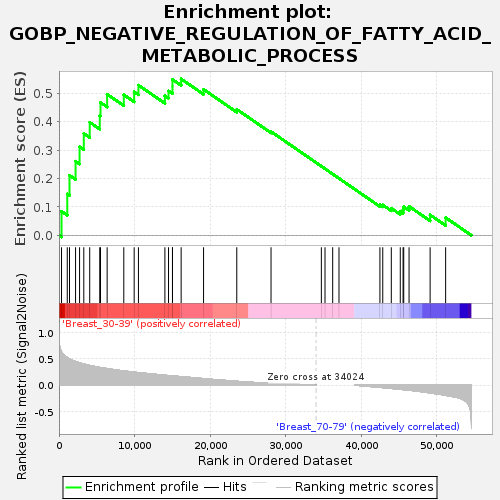
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Breast.Breast.cls #Breast_30-39_versus_Breast_70-79.Breast.cls #Breast_30-39_versus_Breast_70-79_repos |
| Phenotype | Breast.cls#Breast_30-39_versus_Breast_70-79_repos |
| Upregulated in class | Breast_30-39 |
| GeneSet | GOBP_NEGATIVE_REGULATION_OF_FATTY_ACID_METABOLIC_PROCESS |
| Enrichment Score (ES) | 0.5510812 |
| Normalized Enrichment Score (NES) | 1.6267779 |
| Nominal p-value | 0.0051107327 |
| FDR q-value | 0.23377857 |
| FWER p-Value | 0.856 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | APPL2 | NA | 333 | 0.629 | 0.0845 | Yes |
| 2 | BRCA1 | NA | 1094 | 0.522 | 0.1457 | Yes |
| 3 | AKT1 | NA | 1390 | 0.496 | 0.2116 | Yes |
| 4 | ERLIN2 | NA | 2192 | 0.446 | 0.2612 | Yes |
| 5 | SIRT1 | NA | 2723 | 0.421 | 0.3120 | Yes |
| 6 | NCOR1 | NA | 3282 | 0.399 | 0.3592 | Yes |
| 7 | WDTC1 | NA | 4066 | 0.372 | 0.3984 | Yes |
| 8 | INS | NA | 5395 | 0.335 | 0.4222 | Yes |
| 9 | SLC22A13 | NA | 5494 | 0.332 | 0.4682 | Yes |
| 10 | ETFBKMT | NA | 6372 | 0.312 | 0.4971 | Yes |
| 11 | ERLIN1 | NA | 8577 | 0.269 | 0.4953 | Yes |
| 12 | ACADL | NA | 9951 | 0.245 | 0.5054 | Yes |
| 13 | ACADVL | NA | 10516 | 0.236 | 0.5291 | Yes |
| 14 | PLIN5 | NA | 14018 | 0.186 | 0.4917 | Yes |
| 15 | SOX9 | NA | 14498 | 0.180 | 0.5089 | Yes |
| 16 | SIRT4 | NA | 15022 | 0.173 | 0.5242 | Yes |
| 17 | MFSD2A | NA | 15023 | 0.173 | 0.5492 | Yes |
| 18 | ACACB | NA | 16170 | 0.159 | 0.5511 | Yes |
| 19 | CNR1 | NA | 19137 | 0.122 | 0.5143 | No |
| 20 | MIR132 | NA | 23534 | 0.072 | 0.4440 | No |
| 21 | MIR548P | NA | 28068 | 0.032 | 0.3655 | No |
| 22 | MIR33A | NA | 34731 | 0.000 | 0.2434 | No |
| 23 | MIR204 | NA | 35214 | 0.000 | 0.2346 | No |
| 24 | MIR30C1 | NA | 36230 | 0.000 | 0.2160 | No |
| 25 | MIR766 | NA | 37065 | 0.000 | 0.2007 | No |
| 26 | CYP7A1 | NA | 42483 | -0.042 | 0.1074 | No |
| 27 | DGAT2 | NA | 42859 | -0.047 | 0.1072 | No |
| 28 | PIBF1 | NA | 43989 | -0.062 | 0.0954 | No |
| 29 | INSIG1 | NA | 45178 | -0.079 | 0.0850 | No |
| 30 | UGT1A8 | NA | 45548 | -0.084 | 0.0904 | No |
| 31 | APOC1 | NA | 45635 | -0.086 | 0.1011 | No |
| 32 | CEACAM1 | NA | 46340 | -0.097 | 0.1021 | No |
| 33 | APOC3 | NA | 49129 | -0.147 | 0.0722 | No |
| 34 | TRIB3 | NA | 51182 | -0.194 | 0.0625 | No |