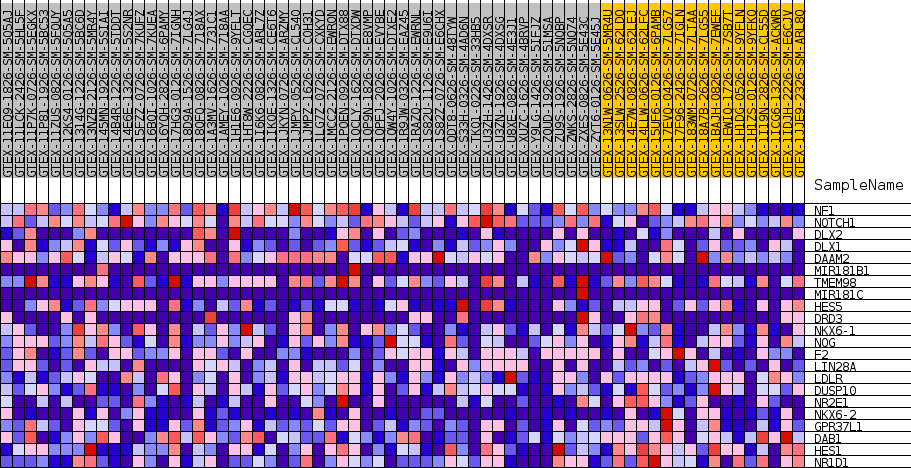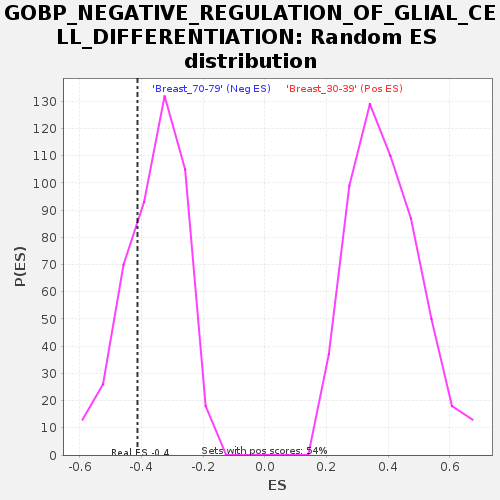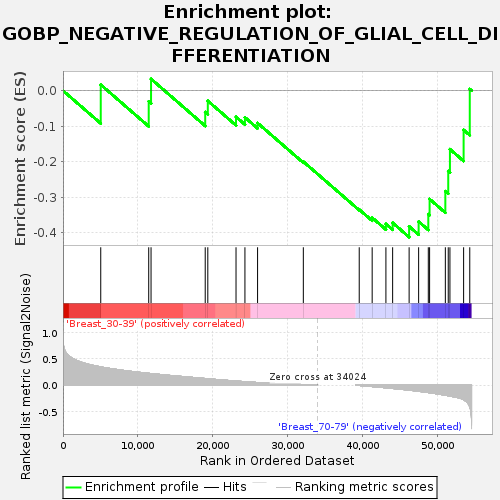
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Breast.Breast.cls #Breast_30-39_versus_Breast_70-79.Breast.cls #Breast_30-39_versus_Breast_70-79_repos |
| Phenotype | Breast.cls#Breast_30-39_versus_Breast_70-79_repos |
| Upregulated in class | Breast_70-79 |
| GeneSet | GOBP_NEGATIVE_REGULATION_OF_GLIAL_CELL_DIFFERENTIATION |
| Enrichment Score (ES) | -0.41253877 |
| Normalized Enrichment Score (NES) | -1.1558778 |
| Nominal p-value | 0.26477024 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.999 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | NF1 | NA | 5043 | 0.344 | 0.0167 | No |
| 2 | NOTCH1 | NA | 11453 | 0.222 | -0.0302 | No |
| 3 | DLX2 | NA | 11764 | 0.218 | 0.0331 | No |
| 4 | DLX1 | NA | 19002 | 0.124 | -0.0603 | No |
| 5 | DAAM2 | NA | 19348 | 0.119 | -0.0288 | No |
| 6 | MIR181B1 | NA | 23118 | 0.076 | -0.0738 | No |
| 7 | TMEM98 | NA | 24308 | 0.064 | -0.0753 | No |
| 8 | MIR181C | NA | 25998 | 0.048 | -0.0912 | No |
| 9 | HES5 | NA | 32111 | 0.010 | -0.1999 | No |
| 10 | DRD3 | NA | 39580 | -0.006 | -0.3349 | No |
| 11 | NKX6-1 | NA | 41309 | -0.027 | -0.3578 | No |
| 12 | NOG | NA | 43145 | -0.050 | -0.3755 | No |
| 13 | F2 | NA | 44044 | -0.062 | -0.3722 | No |
| 14 | LIN28A | NA | 46247 | -0.095 | -0.3824 | Yes |
| 15 | LDLR | NA | 47518 | -0.117 | -0.3687 | Yes |
| 16 | DUSP10 | NA | 48809 | -0.140 | -0.3479 | Yes |
| 17 | NR2E1 | NA | 48983 | -0.144 | -0.3054 | Yes |
| 18 | NKX6-2 | NA | 51086 | -0.191 | -0.2832 | Yes |
| 19 | GPR37L1 | NA | 51474 | -0.201 | -0.2265 | Yes |
| 20 | DAB1 | NA | 51700 | -0.208 | -0.1649 | Yes |
| 21 | HES1 | NA | 53528 | -0.277 | -0.1106 | Yes |
| 22 | NR1D1 | NA | 54350 | -0.410 | 0.0044 | Yes |