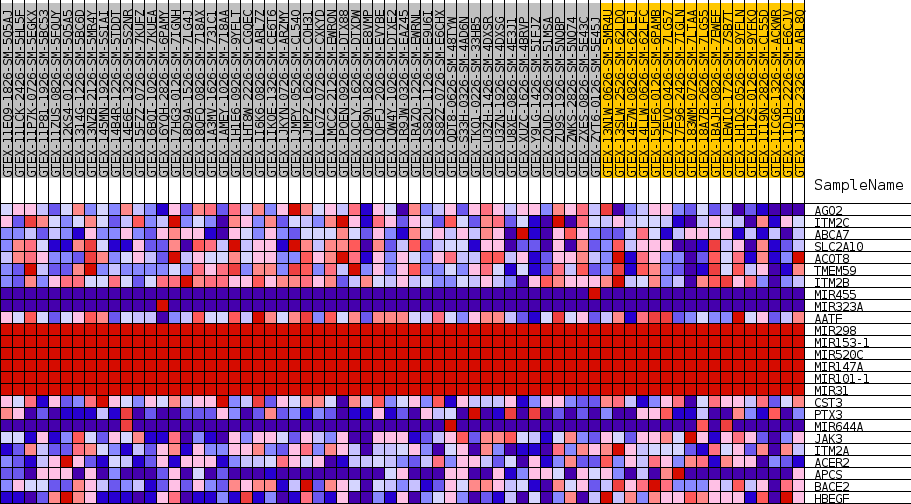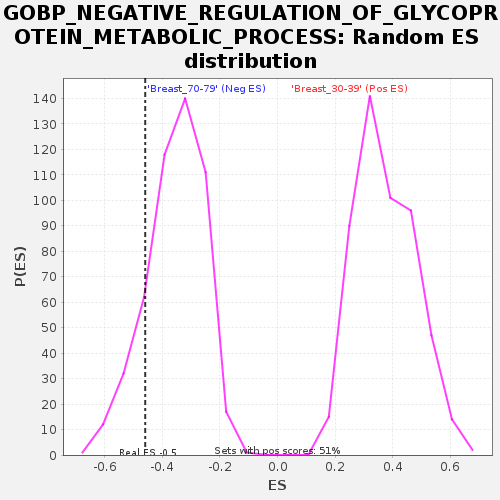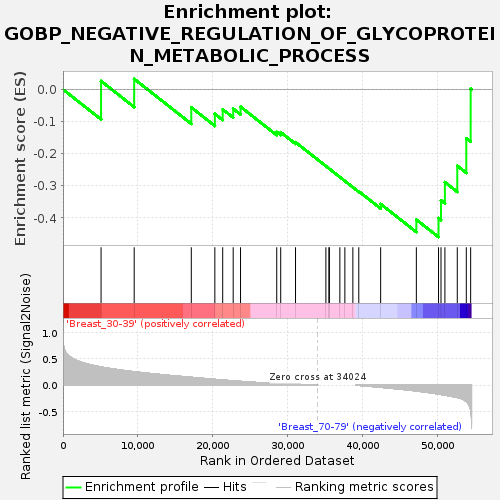
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Breast.Breast.cls #Breast_30-39_versus_Breast_70-79.Breast.cls #Breast_30-39_versus_Breast_70-79_repos |
| Phenotype | Breast.cls#Breast_30-39_versus_Breast_70-79_repos |
| Upregulated in class | Breast_70-79 |
| GeneSet | GOBP_NEGATIVE_REGULATION_OF_GLYCOPROTEIN_METABOLIC_PROCESS |
| Enrichment Score (ES) | -0.45956334 |
| Normalized Enrichment Score (NES) | -1.2932025 |
| Nominal p-value | 0.15789473 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.998 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | AGO2 | NA | 5086 | 0.344 | 0.0261 | No |
| 2 | ITM2C | NA | 9511 | 0.252 | 0.0327 | No |
| 3 | ABCA7 | NA | 17142 | 0.147 | -0.0559 | No |
| 4 | SLC2A10 | NA | 20289 | 0.108 | -0.0761 | No |
| 5 | ACOT8 | NA | 21338 | 0.096 | -0.0621 | No |
| 6 | TMEM59 | NA | 22739 | 0.080 | -0.0600 | No |
| 7 | ITM2B | NA | 23721 | 0.070 | -0.0538 | No |
| 8 | MIR455 | NA | 28558 | 0.029 | -0.1324 | No |
| 9 | MIR323A | NA | 29089 | 0.026 | -0.1332 | No |
| 10 | AATF | NA | 31068 | 0.015 | -0.1642 | No |
| 11 | MIR298 | NA | 35125 | 0.000 | -0.2386 | No |
| 12 | MIR153-1 | NA | 35529 | 0.000 | -0.2460 | No |
| 13 | MIR520C | NA | 35591 | 0.000 | -0.2471 | No |
| 14 | MIR147A | NA | 36991 | 0.000 | -0.2727 | No |
| 15 | MIR101-1 | NA | 37661 | 0.000 | -0.2850 | No |
| 16 | MIR31 | NA | 38730 | 0.000 | -0.3045 | No |
| 17 | CST3 | NA | 39521 | -0.005 | -0.3172 | No |
| 18 | PTX3 | NA | 42442 | -0.041 | -0.3565 | No |
| 19 | MIR644A | NA | 47212 | -0.111 | -0.4053 | Yes |
| 20 | JAK3 | NA | 50175 | -0.170 | -0.4004 | Yes |
| 21 | ITM2A | NA | 50506 | -0.177 | -0.3450 | Yes |
| 22 | ACER2 | NA | 51027 | -0.190 | -0.2886 | Yes |
| 23 | APCS | NA | 52681 | -0.234 | -0.2377 | Yes |
| 24 | BACE2 | NA | 53887 | -0.310 | -0.1521 | Yes |
| 25 | HBEGF | NA | 54468 | -0.475 | 0.0023 | Yes |