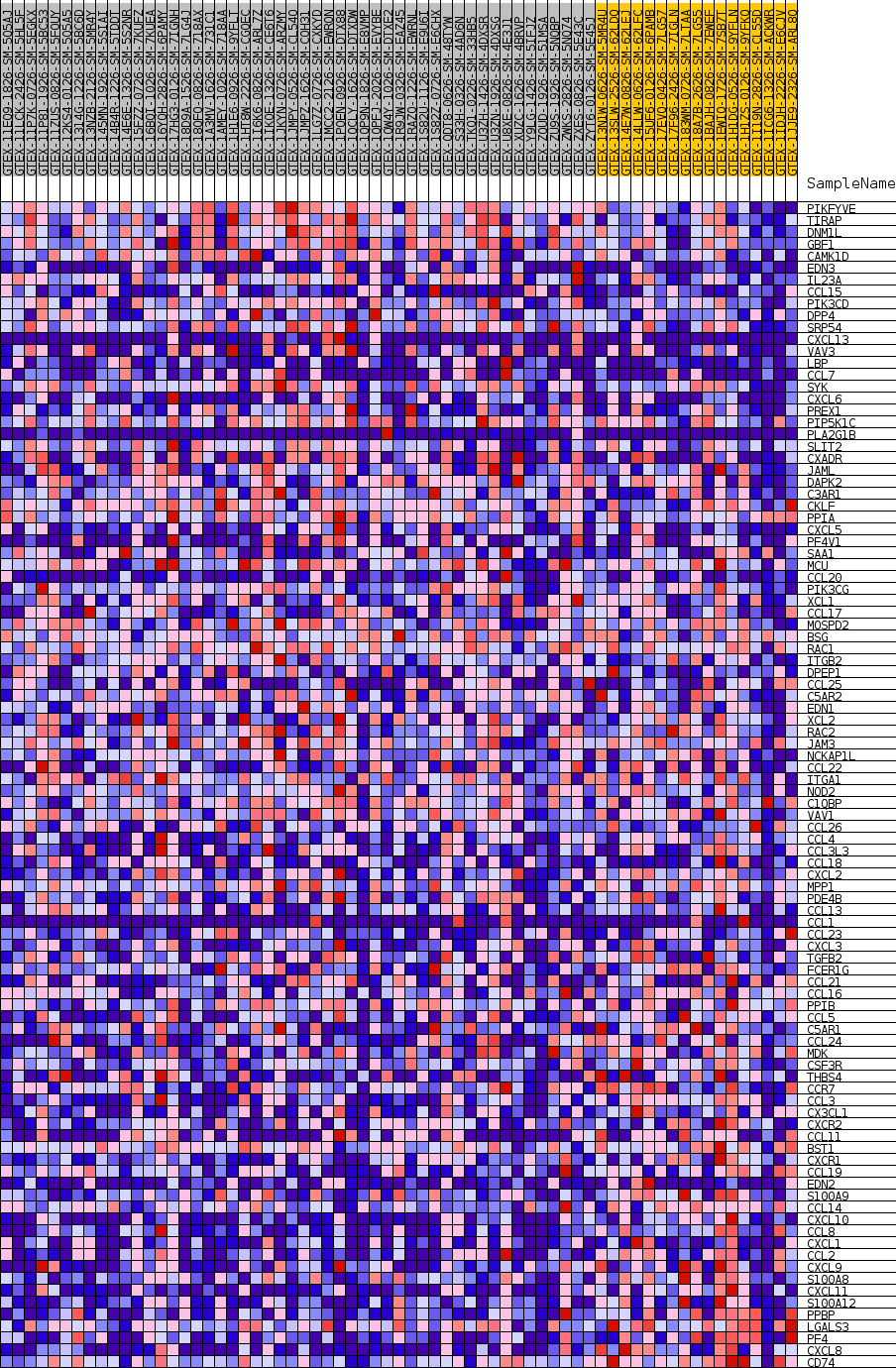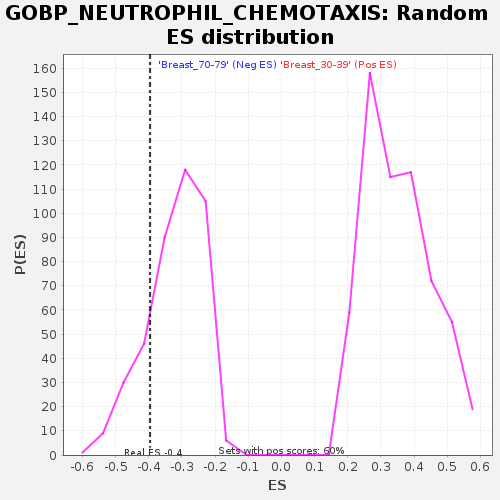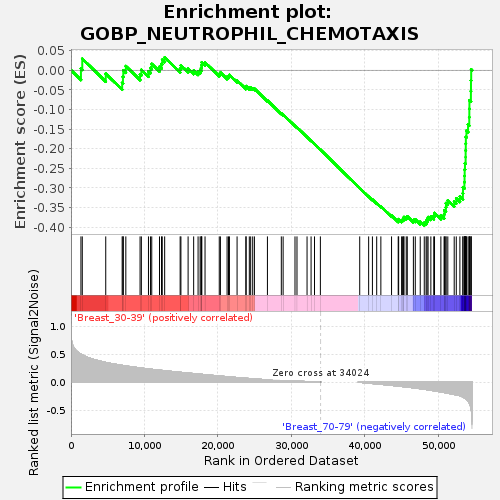
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Breast.Breast.cls #Breast_30-39_versus_Breast_70-79.Breast.cls #Breast_30-39_versus_Breast_70-79_repos |
| Phenotype | Breast.cls#Breast_30-39_versus_Breast_70-79_repos |
| Upregulated in class | Breast_70-79 |
| GeneSet | GOBP_NEUTROPHIL_CHEMOTAXIS |
| Enrichment Score (ES) | -0.396115 |
| Normalized Enrichment Score (NES) | -1.2333814 |
| Nominal p-value | 0.17283951 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.998 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | PIKFYVE | NA | 1353 | 0.499 | 0.0043 | No |
| 2 | TIRAP | NA | 1536 | 0.486 | 0.0294 | No |
| 3 | DNM1L | NA | 4730 | 0.353 | -0.0086 | No |
| 4 | GBF1 | NA | 6966 | 0.300 | -0.0321 | No |
| 5 | CAMK1D | NA | 7072 | 0.297 | -0.0167 | No |
| 6 | EDN3 | NA | 7121 | 0.297 | -0.0002 | No |
| 7 | IL23A | NA | 7465 | 0.290 | 0.0104 | No |
| 8 | CCL15 | NA | 9403 | 0.254 | -0.0103 | No |
| 9 | PIK3CD | NA | 9579 | 0.251 | 0.0012 | No |
| 10 | DPP4 | NA | 10547 | 0.236 | -0.0028 | No |
| 11 | SRP54 | NA | 10820 | 0.231 | 0.0057 | No |
| 12 | CXCL13 | NA | 10995 | 0.229 | 0.0159 | No |
| 13 | VAV3 | NA | 12048 | 0.214 | 0.0091 | No |
| 14 | LBP | NA | 12322 | 0.210 | 0.0164 | No |
| 15 | CCL7 | NA | 12422 | 0.209 | 0.0268 | No |
| 16 | SYK | NA | 12753 | 0.205 | 0.0327 | No |
| 17 | CXCL6 | NA | 14860 | 0.176 | 0.0043 | No |
| 18 | PREX1 | NA | 14965 | 0.174 | 0.0125 | No |
| 19 | PIP5K1C | NA | 15944 | 0.162 | 0.0041 | No |
| 20 | PLA2G1B | NA | 16700 | 0.153 | -0.0009 | No |
| 21 | SLIT2 | NA | 17302 | 0.145 | -0.0034 | No |
| 22 | CXADR | NA | 17582 | 0.142 | -0.0002 | No |
| 23 | JAML | NA | 17712 | 0.140 | 0.0056 | No |
| 24 | DAPK2 | NA | 17775 | 0.139 | 0.0126 | No |
| 25 | C3AR1 | NA | 17807 | 0.139 | 0.0202 | No |
| 26 | CKLF | NA | 18249 | 0.133 | 0.0199 | No |
| 27 | PPIA | NA | 20186 | 0.109 | -0.0093 | No |
| 28 | CXCL5 | NA | 20343 | 0.107 | -0.0059 | No |
| 29 | PF4V1 | NA | 21251 | 0.097 | -0.0169 | No |
| 30 | SAA1 | NA | 21445 | 0.094 | -0.0149 | No |
| 31 | MCU | NA | 21572 | 0.093 | -0.0118 | No |
| 32 | CCL20 | NA | 22612 | 0.081 | -0.0261 | No |
| 33 | PIK3CG | NA | 23809 | 0.069 | -0.0440 | No |
| 34 | XCL1 | NA | 23843 | 0.068 | -0.0406 | No |
| 35 | CCL17 | NA | 24278 | 0.064 | -0.0448 | No |
| 36 | MOSPD2 | NA | 24464 | 0.063 | -0.0446 | No |
| 37 | BSG | NA | 24740 | 0.060 | -0.0461 | No |
| 38 | RAC1 | NA | 24971 | 0.058 | -0.0470 | No |
| 39 | ITGB2 | NA | 26744 | 0.041 | -0.0771 | No |
| 40 | DPEP1 | NA | 28635 | 0.028 | -0.1101 | No |
| 41 | CCL25 | NA | 28881 | 0.027 | -0.1130 | No |
| 42 | C5AR2 | NA | 30497 | 0.018 | -0.1416 | No |
| 43 | EDN1 | NA | 30768 | 0.016 | -0.1456 | No |
| 44 | XCL2 | NA | 32139 | 0.010 | -0.1702 | No |
| 45 | RAC2 | NA | 32687 | 0.008 | -0.1798 | No |
| 46 | JAM3 | NA | 33154 | 0.006 | -0.1879 | No |
| 47 | NCKAP1L | NA | 33953 | 0.001 | -0.2025 | No |
| 48 | CCL22 | NA | 39310 | -0.003 | -0.3007 | No |
| 49 | ITGA1 | NA | 40526 | -0.018 | -0.3219 | No |
| 50 | NOD2 | NA | 41039 | -0.024 | -0.3299 | No |
| 51 | C1QBP | NA | 41621 | -0.031 | -0.3387 | No |
| 52 | VAV1 | NA | 42188 | -0.038 | -0.3469 | No |
| 53 | CCL26 | NA | 43644 | -0.057 | -0.3703 | No |
| 54 | CCL4 | NA | 44550 | -0.070 | -0.3828 | No |
| 55 | CCL3L3 | NA | 44612 | -0.071 | -0.3798 | No |
| 56 | CCL18 | NA | 45013 | -0.077 | -0.3827 | No |
| 57 | CXCL2 | NA | 45149 | -0.078 | -0.3806 | No |
| 58 | MPP1 | NA | 45288 | -0.080 | -0.3784 | No |
| 59 | PDE4B | NA | 45330 | -0.081 | -0.3744 | No |
| 60 | CCL13 | NA | 45642 | -0.086 | -0.3751 | No |
| 61 | CCL1 | NA | 45784 | -0.088 | -0.3726 | No |
| 62 | CCL23 | NA | 46617 | -0.101 | -0.3820 | No |
| 63 | CXCL3 | NA | 46874 | -0.105 | -0.3805 | No |
| 64 | TGFB2 | NA | 47549 | -0.117 | -0.3861 | No |
| 65 | FCER1G | NA | 48098 | -0.127 | -0.3887 | Yes |
| 66 | CCL21 | NA | 48347 | -0.132 | -0.3855 | Yes |
| 67 | CCL16 | NA | 48475 | -0.135 | -0.3800 | Yes |
| 68 | PPIB | NA | 48616 | -0.137 | -0.3746 | Yes |
| 69 | CCL5 | NA | 48995 | -0.145 | -0.3731 | Yes |
| 70 | C5AR1 | NA | 49391 | -0.153 | -0.3714 | Yes |
| 71 | CCL24 | NA | 49494 | -0.155 | -0.3642 | Yes |
| 72 | MDK | NA | 50359 | -0.174 | -0.3699 | Yes |
| 73 | CSF3R | NA | 50803 | -0.184 | -0.3672 | Yes |
| 74 | THBS4 | NA | 50843 | -0.185 | -0.3571 | Yes |
| 75 | CCR7 | NA | 51037 | -0.190 | -0.3496 | Yes |
| 76 | CCL3 | NA | 51058 | -0.191 | -0.3388 | Yes |
| 77 | CX3CL1 | NA | 51304 | -0.197 | -0.3318 | Yes |
| 78 | CXCR2 | NA | 52174 | -0.220 | -0.3349 | Yes |
| 79 | CCL11 | NA | 52457 | -0.227 | -0.3268 | Yes |
| 80 | BST1 | NA | 52949 | -0.243 | -0.3216 | Yes |
| 81 | CXCR1 | NA | 53355 | -0.265 | -0.3136 | Yes |
| 82 | CCL19 | NA | 53370 | -0.266 | -0.2983 | Yes |
| 83 | EDN2 | NA | 53563 | -0.279 | -0.2855 | Yes |
| 84 | S100A9 | NA | 53584 | -0.282 | -0.2694 | Yes |
| 85 | CCL14 | NA | 53600 | -0.283 | -0.2532 | Yes |
| 86 | CXCL10 | NA | 53640 | -0.286 | -0.2372 | Yes |
| 87 | CCL8 | NA | 53728 | -0.293 | -0.2216 | Yes |
| 88 | CXCL1 | NA | 53731 | -0.294 | -0.2045 | Yes |
| 89 | CCL2 | NA | 53749 | -0.296 | -0.1875 | Yes |
| 90 | CXCL9 | NA | 53759 | -0.297 | -0.1703 | Yes |
| 91 | S100A8 | NA | 53840 | -0.305 | -0.1540 | Yes |
| 92 | CXCL11 | NA | 54059 | -0.334 | -0.1384 | Yes |
| 93 | S100A12 | NA | 54225 | -0.369 | -0.1199 | Yes |
| 94 | PPBP | NA | 54235 | -0.372 | -0.0984 | Yes |
| 95 | LGALS3 | NA | 54248 | -0.375 | -0.0767 | Yes |
| 96 | PF4 | NA | 54448 | -0.463 | -0.0533 | Yes |
| 97 | CXCL8 | NA | 54458 | -0.470 | -0.0260 | Yes |
| 98 | CD74 | NA | 54486 | -0.487 | 0.0019 | Yes |