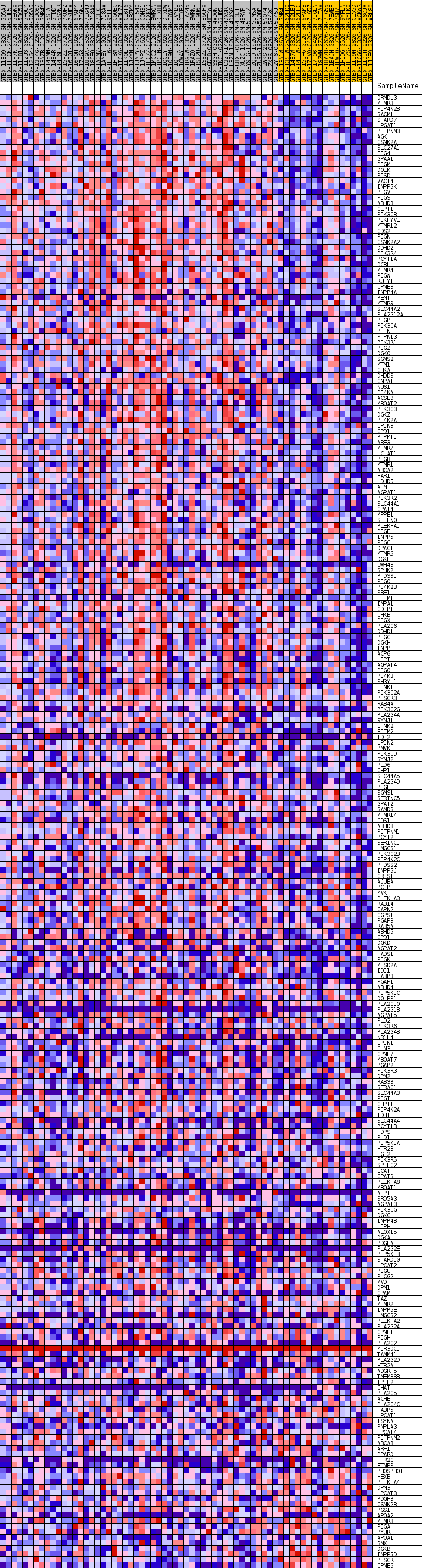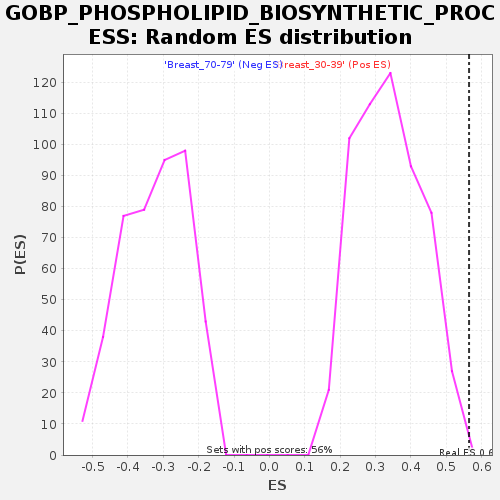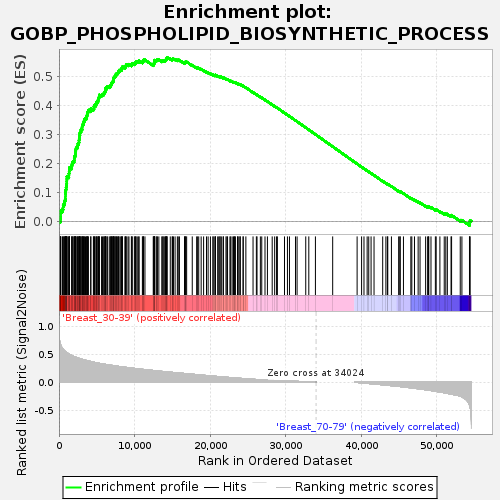
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Breast.Breast.cls #Breast_30-39_versus_Breast_70-79.Breast.cls #Breast_30-39_versus_Breast_70-79_repos |
| Phenotype | Breast.cls#Breast_30-39_versus_Breast_70-79_repos |
| Upregulated in class | Breast_30-39 |
| GeneSet | GOBP_PHOSPHOLIPID_BIOSYNTHETIC_PROCESS |
| Enrichment Score (ES) | 0.5647453 |
| Normalized Enrichment Score (NES) | 1.6718 |
| Nominal p-value | 0.0035778175 |
| FDR q-value | 0.27147382 |
| FWER p-Value | 0.749 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | ORMDL3 | NA | 183 | 0.683 | 0.0070 | Yes |
| 2 | MTMR3 | NA | 209 | 0.668 | 0.0166 | Yes |
| 3 | PIP4K2B | NA | 221 | 0.663 | 0.0264 | Yes |
| 4 | SACM1L | NA | 227 | 0.660 | 0.0363 | Yes |
| 5 | STARD7 | NA | 457 | 0.603 | 0.0412 | Yes |
| 6 | LPGAT1 | NA | 546 | 0.586 | 0.0484 | Yes |
| 7 | PITPNM3 | NA | 558 | 0.584 | 0.0570 | Yes |
| 8 | AGK | NA | 698 | 0.562 | 0.0630 | Yes |
| 9 | CSNK2A1 | NA | 759 | 0.556 | 0.0703 | Yes |
| 10 | SLC27A1 | NA | 819 | 0.549 | 0.0775 | Yes |
| 11 | FIG4 | NA | 851 | 0.545 | 0.0852 | Yes |
| 12 | GPAA1 | NA | 854 | 0.545 | 0.0934 | Yes |
| 13 | PIGM | NA | 877 | 0.543 | 0.1012 | Yes |
| 14 | DOLK | NA | 894 | 0.542 | 0.1090 | Yes |
| 15 | PISD | NA | 944 | 0.536 | 0.1162 | Yes |
| 16 | VAC14 | NA | 970 | 0.533 | 0.1238 | Yes |
| 17 | INPP5K | NA | 1017 | 0.528 | 0.1310 | Yes |
| 18 | PIGV | NA | 1029 | 0.528 | 0.1387 | Yes |
| 19 | PIGS | NA | 1039 | 0.526 | 0.1465 | Yes |
| 20 | ABHD3 | NA | 1049 | 0.525 | 0.1543 | Yes |
| 21 | CEPT1 | NA | 1300 | 0.503 | 0.1573 | Yes |
| 22 | PIK3CB | NA | 1307 | 0.502 | 0.1648 | Yes |
| 23 | PIKFYVE | NA | 1353 | 0.499 | 0.1715 | Yes |
| 24 | MTMR12 | NA | 1368 | 0.497 | 0.1787 | Yes |
| 25 | CDS2 | NA | 1391 | 0.496 | 0.1858 | Yes |
| 26 | PIGN | NA | 1685 | 0.477 | 0.1876 | Yes |
| 27 | CSNK2A2 | NA | 1703 | 0.476 | 0.1945 | Yes |
| 28 | DDHD2 | NA | 1743 | 0.473 | 0.2009 | Yes |
| 29 | PIK3R4 | NA | 1840 | 0.466 | 0.2062 | Yes |
| 30 | PCYT1A | NA | 1993 | 0.456 | 0.2103 | Yes |
| 31 | OCRL | NA | 2023 | 0.454 | 0.2166 | Yes |
| 32 | MTMR4 | NA | 2037 | 0.454 | 0.2233 | Yes |
| 33 | PIGW | NA | 2130 | 0.449 | 0.2283 | Yes |
| 34 | RUFY1 | NA | 2161 | 0.448 | 0.2346 | Yes |
| 35 | CPNE3 | NA | 2172 | 0.448 | 0.2411 | Yes |
| 36 | INPP4A | NA | 2217 | 0.445 | 0.2471 | Yes |
| 37 | PEMT | NA | 2261 | 0.442 | 0.2530 | Yes |
| 38 | MTMR9 | NA | 2410 | 0.435 | 0.2568 | Yes |
| 39 | SLC44A2 | NA | 2472 | 0.432 | 0.2622 | Yes |
| 40 | PLA2G12A | NA | 2515 | 0.430 | 0.2679 | Yes |
| 41 | PIGP | NA | 2621 | 0.426 | 0.2724 | Yes |
| 42 | PIK3CA | NA | 2652 | 0.424 | 0.2783 | Yes |
| 43 | PTEN | NA | 2673 | 0.423 | 0.2843 | Yes |
| 44 | PTPN13 | NA | 2705 | 0.422 | 0.2901 | Yes |
| 45 | PIK3R1 | NA | 2739 | 0.420 | 0.2959 | Yes |
| 46 | PIGZ | NA | 2762 | 0.419 | 0.3018 | Yes |
| 47 | DGKQ | NA | 2788 | 0.418 | 0.3076 | Yes |
| 48 | SGMS2 | NA | 2903 | 0.413 | 0.3118 | Yes |
| 49 | MTM1 | NA | 2916 | 0.412 | 0.3178 | Yes |
| 50 | CHKA | NA | 3068 | 0.406 | 0.3212 | Yes |
| 51 | DHDDS | NA | 3071 | 0.406 | 0.3273 | Yes |
| 52 | GNPAT | NA | 3103 | 0.405 | 0.3328 | Yes |
| 53 | NUS1 | NA | 3201 | 0.402 | 0.3371 | Yes |
| 54 | PI4KA | NA | 3246 | 0.400 | 0.3423 | Yes |
| 55 | ACSL3 | NA | 3331 | 0.397 | 0.3468 | Yes |
| 56 | MBOAT2 | NA | 3362 | 0.396 | 0.3522 | Yes |
| 57 | PIK3C3 | NA | 3507 | 0.391 | 0.3555 | Yes |
| 58 | DGKZ | NA | 3668 | 0.385 | 0.3583 | Yes |
| 59 | PI4K2A | NA | 3675 | 0.385 | 0.3640 | Yes |
| 60 | LPIN3 | NA | 3734 | 0.382 | 0.3687 | Yes |
| 61 | GPD1L | NA | 3756 | 0.382 | 0.3741 | Yes |
| 62 | PTPMT1 | NA | 3833 | 0.379 | 0.3785 | Yes |
| 63 | ARF3 | NA | 3888 | 0.377 | 0.3832 | Yes |
| 64 | MTMR7 | NA | 4199 | 0.368 | 0.3830 | Yes |
| 65 | LCLAT1 | NA | 4217 | 0.368 | 0.3883 | Yes |
| 66 | PIGB | NA | 4552 | 0.358 | 0.3875 | Yes |
| 67 | MTMR1 | NA | 4630 | 0.356 | 0.3915 | Yes |
| 68 | ABCA2 | NA | 4634 | 0.355 | 0.3968 | Yes |
| 69 | FAR1 | NA | 4703 | 0.353 | 0.4009 | Yes |
| 70 | HDHD5 | NA | 4894 | 0.348 | 0.4026 | Yes |
| 71 | ATM | NA | 4915 | 0.348 | 0.4075 | Yes |
| 72 | AGPAT1 | NA | 4979 | 0.346 | 0.4116 | Yes |
| 73 | PIK3R2 | NA | 5143 | 0.342 | 0.4137 | Yes |
| 74 | SLC44A1 | NA | 5167 | 0.341 | 0.4185 | Yes |
| 75 | GPAT4 | NA | 5223 | 0.339 | 0.4226 | Yes |
| 76 | MPPE1 | NA | 5286 | 0.338 | 0.4265 | Yes |
| 77 | SELENOI | NA | 5319 | 0.337 | 0.4310 | Yes |
| 78 | PLEKHA1 | NA | 5339 | 0.336 | 0.4358 | Yes |
| 79 | PIGF | NA | 5633 | 0.329 | 0.4354 | Yes |
| 80 | INPP5F | NA | 5763 | 0.326 | 0.4379 | Yes |
| 81 | PIGC | NA | 5925 | 0.322 | 0.4398 | Yes |
| 82 | DPAGT1 | NA | 5979 | 0.321 | 0.4437 | Yes |
| 83 | MTMR6 | NA | 6066 | 0.319 | 0.4469 | Yes |
| 84 | DGKE | NA | 6163 | 0.317 | 0.4499 | Yes |
| 85 | CWH43 | NA | 6171 | 0.317 | 0.4546 | Yes |
| 86 | SPHK2 | NA | 6175 | 0.317 | 0.4593 | Yes |
| 87 | PTDSS1 | NA | 6348 | 0.313 | 0.4609 | Yes |
| 88 | PIGQ | NA | 6397 | 0.312 | 0.4647 | Yes |
| 89 | PI4K2B | NA | 6648 | 0.306 | 0.4647 | Yes |
| 90 | SBF1 | NA | 6786 | 0.303 | 0.4668 | Yes |
| 91 | FITM1 | NA | 6859 | 0.302 | 0.4700 | Yes |
| 92 | IMPA1 | NA | 6915 | 0.301 | 0.4736 | Yes |
| 93 | CDIPT | NA | 6957 | 0.300 | 0.4774 | Yes |
| 94 | CHKB | NA | 7051 | 0.298 | 0.4801 | Yes |
| 95 | PIGX | NA | 7148 | 0.296 | 0.4828 | Yes |
| 96 | PLA2G6 | NA | 7179 | 0.296 | 0.4868 | Yes |
| 97 | DDHD1 | NA | 7190 | 0.295 | 0.4910 | Yes |
| 98 | PIGG | NA | 7224 | 0.295 | 0.4949 | Yes |
| 99 | DGKH | NA | 7314 | 0.293 | 0.4977 | Yes |
| 100 | INPPL1 | NA | 7421 | 0.290 | 0.5001 | Yes |
| 101 | ACP6 | NA | 7440 | 0.290 | 0.5042 | Yes |
| 102 | LIPI | NA | 7580 | 0.287 | 0.5059 | Yes |
| 103 | AGPAT4 | NA | 7620 | 0.287 | 0.5096 | Yes |
| 104 | PIGO | NA | 7764 | 0.284 | 0.5112 | Yes |
| 105 | PI4KB | NA | 7834 | 0.282 | 0.5142 | Yes |
| 106 | SH3YL1 | NA | 7864 | 0.282 | 0.5179 | Yes |
| 107 | ETNK1 | NA | 7952 | 0.280 | 0.5206 | Yes |
| 108 | PIK3C2A | NA | 8112 | 0.277 | 0.5218 | Yes |
| 109 | PLSCR3 | NA | 8268 | 0.274 | 0.5231 | Yes |
| 110 | RAB4A | NA | 8331 | 0.273 | 0.5261 | Yes |
| 111 | PIK3C2G | NA | 8342 | 0.273 | 0.5300 | Yes |
| 112 | PLA2G4A | NA | 8399 | 0.272 | 0.5331 | Yes |
| 113 | SYNJ1 | NA | 8722 | 0.266 | 0.5312 | Yes |
| 114 | ETNK2 | NA | 8802 | 0.265 | 0.5337 | Yes |
| 115 | FITM2 | NA | 8853 | 0.264 | 0.5368 | Yes |
| 116 | IDI2 | NA | 8901 | 0.263 | 0.5399 | Yes |
| 117 | LPIN2 | NA | 9132 | 0.259 | 0.5396 | Yes |
| 118 | PMVK | NA | 9260 | 0.257 | 0.5411 | Yes |
| 119 | PIK3CD | NA | 9579 | 0.251 | 0.5391 | Yes |
| 120 | SYNJ2 | NA | 9642 | 0.250 | 0.5417 | Yes |
| 121 | PLD6 | NA | 9694 | 0.249 | 0.5445 | Yes |
| 122 | CHP1 | NA | 9977 | 0.245 | 0.5431 | Yes |
| 123 | SLC44A5 | NA | 10080 | 0.243 | 0.5449 | Yes |
| 124 | PLA2G4D | NA | 10114 | 0.243 | 0.5479 | Yes |
| 125 | PIGL | NA | 10207 | 0.241 | 0.5499 | Yes |
| 126 | SGMS1 | NA | 10358 | 0.239 | 0.5507 | Yes |
| 127 | SERINC5 | NA | 10552 | 0.236 | 0.5507 | Yes |
| 128 | GPAT2 | NA | 10600 | 0.235 | 0.5534 | Yes |
| 129 | SAMD8 | NA | 11037 | 0.229 | 0.5488 | Yes |
| 130 | MTMR14 | NA | 11106 | 0.227 | 0.5510 | Yes |
| 131 | CDS1 | NA | 11169 | 0.227 | 0.5533 | Yes |
| 132 | ABHD8 | NA | 11193 | 0.226 | 0.5563 | Yes |
| 133 | PITPNM1 | NA | 11378 | 0.223 | 0.5563 | Yes |
| 134 | PCYT2 | NA | 12476 | 0.208 | 0.5392 | Yes |
| 135 | SERINC1 | NA | 12545 | 0.207 | 0.5411 | Yes |
| 136 | HMGCS1 | NA | 12572 | 0.207 | 0.5437 | Yes |
| 137 | PIK3C2B | NA | 12594 | 0.206 | 0.5465 | Yes |
| 138 | PIP4K2C | NA | 12621 | 0.206 | 0.5491 | Yes |
| 139 | PTDSS2 | NA | 12630 | 0.206 | 0.5521 | Yes |
| 140 | INPP5J | NA | 12632 | 0.206 | 0.5552 | Yes |
| 141 | CRLS1 | NA | 12850 | 0.203 | 0.5542 | Yes |
| 142 | AJUBA | NA | 12946 | 0.201 | 0.5555 | Yes |
| 143 | PCTP | NA | 13027 | 0.200 | 0.5571 | Yes |
| 144 | MVK | NA | 13218 | 0.198 | 0.5566 | Yes |
| 145 | PLEKHA3 | NA | 13606 | 0.192 | 0.5524 | Yes |
| 146 | RAB14 | NA | 13732 | 0.190 | 0.5529 | Yes |
| 147 | CAPN2 | NA | 13794 | 0.190 | 0.5547 | Yes |
| 148 | GGPS1 | NA | 13940 | 0.187 | 0.5548 | Yes |
| 149 | PGAP3 | NA | 14096 | 0.185 | 0.5548 | Yes |
| 150 | RAB5A | NA | 14128 | 0.185 | 0.5570 | Yes |
| 151 | ABHD5 | NA | 14212 | 0.184 | 0.5583 | Yes |
| 152 | GPD1 | NA | 14218 | 0.184 | 0.5609 | Yes |
| 153 | DGKD | NA | 14282 | 0.183 | 0.5625 | Yes |
| 154 | AGPAT2 | NA | 14313 | 0.182 | 0.5647 | Yes |
| 155 | FADS1 | NA | 14730 | 0.177 | 0.5598 | No |
| 156 | PIGK | NA | 14980 | 0.174 | 0.5578 | No |
| 157 | MFSD2A | NA | 15023 | 0.173 | 0.5597 | No |
| 158 | IDI1 | NA | 15163 | 0.172 | 0.5597 | No |
| 159 | FABP3 | NA | 15369 | 0.169 | 0.5585 | No |
| 160 | PGAP1 | NA | 15667 | 0.166 | 0.5555 | No |
| 161 | ABHD4 | NA | 15785 | 0.164 | 0.5558 | No |
| 162 | PIP5K1C | NA | 15944 | 0.162 | 0.5554 | No |
| 163 | DOLPP1 | NA | 16616 | 0.154 | 0.5454 | No |
| 164 | PLA2G10 | NA | 16686 | 0.153 | 0.5464 | No |
| 165 | PLA2G1B | NA | 16700 | 0.153 | 0.5485 | No |
| 166 | AGPAT5 | NA | 16776 | 0.152 | 0.5494 | No |
| 167 | PLD2 | NA | 16898 | 0.150 | 0.5494 | No |
| 168 | PIK3R6 | NA | 17640 | 0.141 | 0.5379 | No |
| 169 | PLA2G4B | NA | 18222 | 0.134 | 0.5292 | No |
| 170 | NR1H4 | NA | 18353 | 0.132 | 0.5289 | No |
| 171 | LPIN1 | NA | 18498 | 0.130 | 0.5282 | No |
| 172 | CLN3 | NA | 18819 | 0.126 | 0.5242 | No |
| 173 | CPNE7 | NA | 19143 | 0.122 | 0.5201 | No |
| 174 | MBOAT7 | NA | 19526 | 0.117 | 0.5148 | No |
| 175 | PGAP2 | NA | 19740 | 0.114 | 0.5126 | No |
| 176 | PIK3R3 | NA | 20047 | 0.111 | 0.5087 | No |
| 177 | DPM2 | NA | 20379 | 0.107 | 0.5042 | No |
| 178 | RAB38 | NA | 20404 | 0.106 | 0.5054 | No |
| 179 | SERAC1 | NA | 20639 | 0.104 | 0.5026 | No |
| 180 | SLC44A3 | NA | 20682 | 0.103 | 0.5034 | No |
| 181 | PIGT | NA | 20709 | 0.103 | 0.5045 | No |
| 182 | CHPT1 | NA | 21006 | 0.099 | 0.5005 | No |
| 183 | PIP4K2A | NA | 21146 | 0.098 | 0.4995 | No |
| 184 | IDH1 | NA | 21302 | 0.096 | 0.4981 | No |
| 185 | SLC44A4 | NA | 21317 | 0.096 | 0.4992 | No |
| 186 | PCYT1B | NA | 21489 | 0.094 | 0.4975 | No |
| 187 | FDPS | NA | 21726 | 0.091 | 0.4945 | No |
| 188 | PLD1 | NA | 21765 | 0.091 | 0.4952 | No |
| 189 | PIP5K1A | NA | 22104 | 0.087 | 0.4903 | No |
| 190 | HTR2B | NA | 22228 | 0.086 | 0.4893 | No |
| 191 | FGF2 | NA | 22312 | 0.085 | 0.4891 | No |
| 192 | PIK3R5 | NA | 22627 | 0.081 | 0.4845 | No |
| 193 | SPTLC2 | NA | 22724 | 0.080 | 0.4840 | No |
| 194 | LCAT | NA | 22918 | 0.078 | 0.4816 | No |
| 195 | GPAT3 | NA | 23098 | 0.076 | 0.4795 | No |
| 196 | PLEKHA8 | NA | 23200 | 0.075 | 0.4787 | No |
| 197 | MBOAT1 | NA | 23251 | 0.075 | 0.4790 | No |
| 198 | ALPI | NA | 23354 | 0.073 | 0.4782 | No |
| 199 | SRD5A3 | NA | 23628 | 0.071 | 0.4742 | No |
| 200 | AGPAT3 | NA | 23670 | 0.070 | 0.4745 | No |
| 201 | PIK3CG | NA | 23809 | 0.069 | 0.4730 | No |
| 202 | DGKG | NA | 23961 | 0.067 | 0.4713 | No |
| 203 | INPP4B | NA | 23997 | 0.067 | 0.4716 | No |
| 204 | LIPH | NA | 24369 | 0.063 | 0.4658 | No |
| 205 | ALOX15 | NA | 24387 | 0.063 | 0.4664 | No |
| 206 | DGKA | NA | 24737 | 0.060 | 0.4609 | No |
| 207 | PDGFA | NA | 25687 | 0.051 | 0.4442 | No |
| 208 | PLA2G2E | NA | 26126 | 0.047 | 0.4368 | No |
| 209 | PIP5K1B | NA | 26213 | 0.046 | 0.4359 | No |
| 210 | STARD10 | NA | 26655 | 0.042 | 0.4285 | No |
| 211 | LPCAT2 | NA | 26841 | 0.041 | 0.4257 | No |
| 212 | PIGU | NA | 27270 | 0.037 | 0.4183 | No |
| 213 | PLCG2 | NA | 27591 | 0.035 | 0.4130 | No |
| 214 | MVD | NA | 28223 | 0.031 | 0.4018 | No |
| 215 | DPM1 | NA | 28550 | 0.029 | 0.3963 | No |
| 216 | GPAM | NA | 28801 | 0.027 | 0.3921 | No |
| 217 | TAZ | NA | 28909 | 0.027 | 0.3905 | No |
| 218 | MTMR2 | NA | 29851 | 0.021 | 0.3735 | No |
| 219 | INPP5E | NA | 30241 | 0.019 | 0.3667 | No |
| 220 | HMGCS2 | NA | 30508 | 0.018 | 0.3620 | No |
| 221 | PLEKHA2 | NA | 31306 | 0.014 | 0.3476 | No |
| 222 | PLA2G2A | NA | 31517 | 0.013 | 0.3439 | No |
| 223 | CPNE1 | NA | 32664 | 0.008 | 0.3229 | No |
| 224 | PIGH | NA | 33069 | 0.007 | 0.3156 | No |
| 225 | PLA2G2F | NA | 33945 | 0.001 | 0.2995 | No |
| 226 | MIR30C1 | NA | 36230 | 0.000 | 0.2575 | No |
| 227 | TAMM41 | NA | 39464 | -0.005 | 0.1980 | No |
| 228 | PLA2G2D | NA | 40056 | -0.012 | 0.1873 | No |
| 229 | HTR2A | NA | 40373 | -0.016 | 0.1817 | No |
| 230 | ADGRF5 | NA | 40792 | -0.022 | 0.1744 | No |
| 231 | TMEM38B | NA | 41004 | -0.024 | 0.1709 | No |
| 232 | TPTE2 | NA | 41312 | -0.027 | 0.1656 | No |
| 233 | CHAT | NA | 41704 | -0.032 | 0.1589 | No |
| 234 | PLA2G5 | NA | 42850 | -0.047 | 0.1385 | No |
| 235 | ACHE | NA | 43273 | -0.052 | 0.1316 | No |
| 236 | PLA2G4C | NA | 43475 | -0.055 | 0.1287 | No |
| 237 | FABP5 | NA | 43523 | -0.055 | 0.1286 | No |
| 238 | LPCAT1 | NA | 44007 | -0.062 | 0.1207 | No |
| 239 | ISYNA1 | NA | 44963 | -0.076 | 0.1043 | No |
| 240 | PNPLA3 | NA | 45113 | -0.078 | 0.1027 | No |
| 241 | LPCAT4 | NA | 45175 | -0.079 | 0.1028 | No |
| 242 | PITPNM2 | NA | 45598 | -0.085 | 0.0963 | No |
| 243 | ABCA8 | NA | 46577 | -0.100 | 0.0798 | No |
| 244 | ARF1 | NA | 46706 | -0.102 | 0.0790 | No |
| 245 | PPARD | NA | 47058 | -0.108 | 0.0742 | No |
| 246 | HTR2C | NA | 47543 | -0.117 | 0.0670 | No |
| 247 | ETNPPL | NA | 47794 | -0.122 | 0.0643 | No |
| 248 | PHOSPHO1 | NA | 48535 | -0.135 | 0.0527 | No |
| 249 | HEXB | NA | 48800 | -0.140 | 0.0499 | No |
| 250 | PLEKHA4 | NA | 48933 | -0.143 | 0.0497 | No |
| 251 | DPM3 | NA | 48935 | -0.143 | 0.0518 | No |
| 252 | LPCAT3 | NA | 49243 | -0.150 | 0.0484 | No |
| 253 | PDGFB | NA | 49822 | -0.162 | 0.0402 | No |
| 254 | CSNK2B | NA | 49931 | -0.165 | 0.0407 | No |
| 255 | PGS1 | NA | 50423 | -0.175 | 0.0344 | No |
| 256 | APOA2 | NA | 50997 | -0.189 | 0.0267 | No |
| 257 | MTMR8 | NA | 51205 | -0.194 | 0.0258 | No |
| 258 | PIGA | NA | 51410 | -0.200 | 0.0251 | No |
| 259 | PYURF | NA | 51900 | -0.212 | 0.0193 | No |
| 260 | APOA1 | NA | 51971 | -0.214 | 0.0212 | No |
| 261 | BMX | NA | 53116 | -0.251 | 0.0039 | No |
| 262 | DGKB | NA | 53337 | -0.263 | 0.0039 | No |
| 263 | INPP5D | NA | 54339 | -0.407 | -0.0084 | No |
| 264 | PLSCR1 | NA | 54364 | -0.417 | -0.0026 | No |
| 265 | CPNE6 | NA | 54424 | -0.444 | 0.0031 | No |