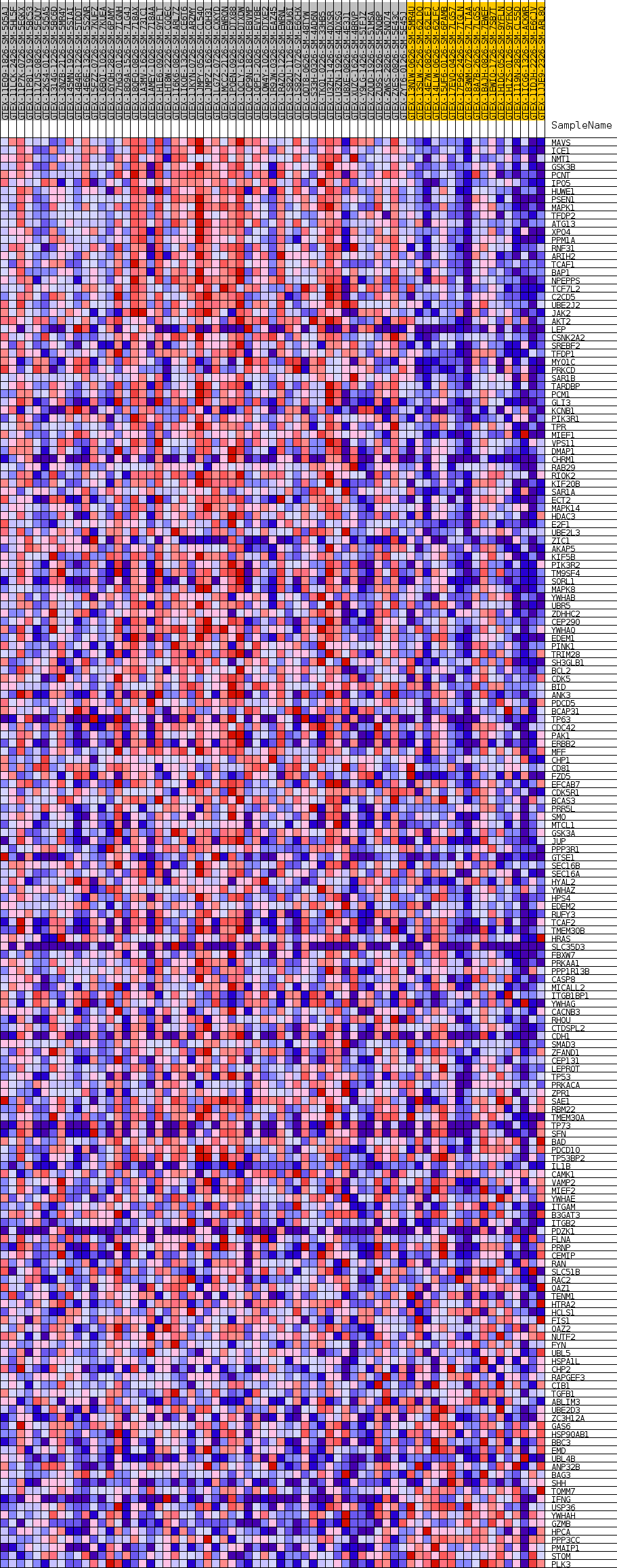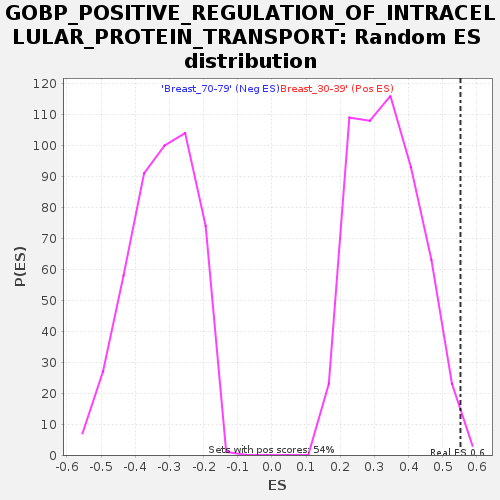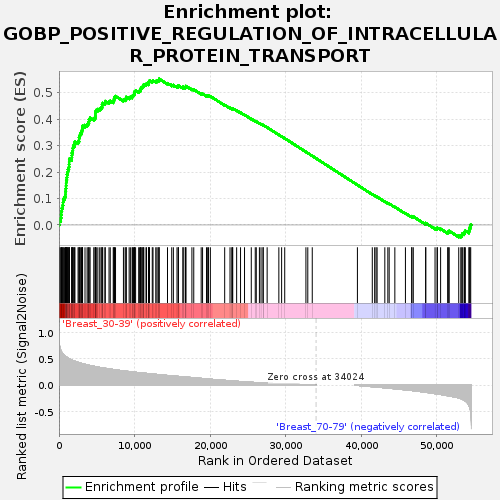
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Breast.Breast.cls #Breast_30-39_versus_Breast_70-79.Breast.cls #Breast_30-39_versus_Breast_70-79_repos |
| Phenotype | Breast.cls#Breast_30-39_versus_Breast_70-79_repos |
| Upregulated in class | Breast_30-39 |
| GeneSet | GOBP_POSITIVE_REGULATION_OF_INTRACELLULAR_PROTEIN_TRANSPORT |
| Enrichment Score (ES) | 0.552057 |
| Normalized Enrichment Score (NES) | 1.6419369 |
| Nominal p-value | 0.00929368 |
| FDR q-value | 0.23753197 |
| FWER p-Value | 0.824 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | MAVS | NA | 41 | 0.780 | 0.0157 | Yes |
| 2 | ICE1 | NA | 234 | 0.658 | 0.0261 | Yes |
| 3 | NMT1 | NA | 276 | 0.648 | 0.0390 | Yes |
| 4 | GSK3B | NA | 325 | 0.632 | 0.0514 | Yes |
| 5 | PCNT | NA | 375 | 0.619 | 0.0636 | Yes |
| 6 | IPO5 | NA | 483 | 0.597 | 0.0742 | Yes |
| 7 | HUWE1 | NA | 540 | 0.587 | 0.0856 | Yes |
| 8 | PSEN1 | NA | 559 | 0.584 | 0.0976 | Yes |
| 9 | MAPK1 | NA | 738 | 0.558 | 0.1061 | Yes |
| 10 | TFDP2 | NA | 857 | 0.545 | 0.1154 | Yes |
| 11 | ATG13 | NA | 866 | 0.544 | 0.1268 | Yes |
| 12 | XPO4 | NA | 904 | 0.541 | 0.1375 | Yes |
| 13 | PPM1A | NA | 935 | 0.536 | 0.1483 | Yes |
| 14 | RNF31 | NA | 950 | 0.535 | 0.1593 | Yes |
| 15 | ARIH2 | NA | 961 | 0.534 | 0.1704 | Yes |
| 16 | TCAF1 | NA | 1041 | 0.526 | 0.1801 | Yes |
| 17 | BAP1 | NA | 1060 | 0.525 | 0.1908 | Yes |
| 18 | NPEPPS | NA | 1145 | 0.517 | 0.2002 | Yes |
| 19 | TCF7L2 | NA | 1189 | 0.513 | 0.2102 | Yes |
| 20 | C2CD5 | NA | 1305 | 0.503 | 0.2187 | Yes |
| 21 | UBE2J2 | NA | 1334 | 0.500 | 0.2288 | Yes |
| 22 | JAK2 | NA | 1350 | 0.499 | 0.2390 | Yes |
| 23 | AKT2 | NA | 1358 | 0.498 | 0.2494 | Yes |
| 24 | LEP | NA | 1684 | 0.477 | 0.2535 | Yes |
| 25 | CSNK2A2 | NA | 1703 | 0.476 | 0.2632 | Yes |
| 26 | SREBF2 | NA | 1718 | 0.475 | 0.2730 | Yes |
| 27 | TFDP1 | NA | 1817 | 0.467 | 0.2810 | Yes |
| 28 | MYO1C | NA | 1832 | 0.466 | 0.2906 | Yes |
| 29 | PRKCD | NA | 1888 | 0.463 | 0.2994 | Yes |
| 30 | SAR1B | NA | 2065 | 0.452 | 0.3057 | Yes |
| 31 | TARDBP | NA | 2102 | 0.450 | 0.3145 | Yes |
| 32 | PCM1 | NA | 2524 | 0.430 | 0.3159 | Yes |
| 33 | GLI3 | NA | 2655 | 0.424 | 0.3224 | Yes |
| 34 | KCNB1 | NA | 2659 | 0.424 | 0.3313 | Yes |
| 35 | PIK3R1 | NA | 2739 | 0.420 | 0.3387 | Yes |
| 36 | TPR | NA | 2817 | 0.417 | 0.3461 | Yes |
| 37 | MIEF1 | NA | 2986 | 0.410 | 0.3517 | Yes |
| 38 | VPS11 | NA | 3042 | 0.408 | 0.3593 | Yes |
| 39 | DMAP1 | NA | 3110 | 0.405 | 0.3666 | Yes |
| 40 | CHRM1 | NA | 3120 | 0.405 | 0.3750 | Yes |
| 41 | RAB29 | NA | 3442 | 0.393 | 0.3774 | Yes |
| 42 | RIOK2 | NA | 3721 | 0.383 | 0.3804 | Yes |
| 43 | KIF20B | NA | 3863 | 0.378 | 0.3857 | Yes |
| 44 | SAR1A | NA | 3957 | 0.375 | 0.3920 | Yes |
| 45 | ECT2 | NA | 3975 | 0.375 | 0.3996 | Yes |
| 46 | MAPK14 | NA | 4118 | 0.371 | 0.4048 | Yes |
| 47 | HDAC3 | NA | 4602 | 0.356 | 0.4034 | Yes |
| 48 | E2F1 | NA | 4780 | 0.351 | 0.4076 | Yes |
| 49 | UBE2L3 | NA | 4811 | 0.350 | 0.4144 | Yes |
| 50 | ZIC1 | NA | 4818 | 0.350 | 0.4217 | Yes |
| 51 | AKAP5 | NA | 4825 | 0.350 | 0.4290 | Yes |
| 52 | KIF5B | NA | 4934 | 0.347 | 0.4343 | Yes |
| 53 | PIK3R2 | NA | 5143 | 0.342 | 0.4377 | Yes |
| 54 | TM9SF4 | NA | 5389 | 0.335 | 0.4403 | Yes |
| 55 | SORL1 | NA | 5570 | 0.331 | 0.4440 | Yes |
| 56 | MAPK8 | NA | 5699 | 0.327 | 0.4485 | Yes |
| 57 | YWHAB | NA | 5706 | 0.327 | 0.4553 | Yes |
| 58 | UBR5 | NA | 5788 | 0.325 | 0.4607 | Yes |
| 59 | ZDHHC2 | NA | 6079 | 0.319 | 0.4621 | Yes |
| 60 | CEP290 | NA | 6124 | 0.318 | 0.4680 | Yes |
| 61 | YWHAQ | NA | 6624 | 0.306 | 0.4653 | Yes |
| 62 | EDEM1 | NA | 6784 | 0.303 | 0.4688 | Yes |
| 63 | PINK1 | NA | 7180 | 0.296 | 0.4678 | Yes |
| 64 | TRIM28 | NA | 7261 | 0.294 | 0.4725 | Yes |
| 65 | SH3GLB1 | NA | 7301 | 0.293 | 0.4780 | Yes |
| 66 | BCL2 | NA | 7349 | 0.292 | 0.4833 | Yes |
| 67 | CDK5 | NA | 7483 | 0.289 | 0.4869 | Yes |
| 68 | BID | NA | 8530 | 0.270 | 0.4734 | Yes |
| 69 | ANK3 | NA | 8751 | 0.266 | 0.4749 | Yes |
| 70 | PDCD5 | NA | 8886 | 0.263 | 0.4780 | Yes |
| 71 | BCAP31 | NA | 8891 | 0.263 | 0.4835 | Yes |
| 72 | TP63 | NA | 9285 | 0.256 | 0.4817 | Yes |
| 73 | CDC42 | NA | 9447 | 0.254 | 0.4841 | Yes |
| 74 | PAK1 | NA | 9677 | 0.250 | 0.4852 | Yes |
| 75 | ERBB2 | NA | 9744 | 0.248 | 0.4892 | Yes |
| 76 | MFF | NA | 9869 | 0.247 | 0.4921 | Yes |
| 77 | CHP1 | NA | 9977 | 0.245 | 0.4953 | Yes |
| 78 | CD81 | NA | 9987 | 0.245 | 0.5003 | Yes |
| 79 | FZD5 | NA | 10015 | 0.244 | 0.5050 | Yes |
| 80 | EFCAB7 | NA | 10141 | 0.242 | 0.5078 | Yes |
| 81 | CDK5R1 | NA | 10549 | 0.236 | 0.5053 | Yes |
| 82 | BCAS3 | NA | 10672 | 0.234 | 0.5080 | Yes |
| 83 | PRR5L | NA | 10803 | 0.232 | 0.5105 | Yes |
| 84 | SMO | NA | 10809 | 0.232 | 0.5153 | Yes |
| 85 | MTCL1 | NA | 10874 | 0.231 | 0.5190 | Yes |
| 86 | GSK3A | NA | 11015 | 0.229 | 0.5212 | Yes |
| 87 | JUP | NA | 11135 | 0.227 | 0.5239 | Yes |
| 88 | PPP3R1 | NA | 11154 | 0.227 | 0.5283 | Yes |
| 89 | GTSE1 | NA | 11229 | 0.226 | 0.5317 | Yes |
| 90 | SEC16B | NA | 11518 | 0.221 | 0.5311 | Yes |
| 91 | SEC16A | NA | 11586 | 0.220 | 0.5345 | Yes |
| 92 | HYAL2 | NA | 11859 | 0.216 | 0.5341 | Yes |
| 93 | YWHAZ | NA | 11892 | 0.216 | 0.5381 | Yes |
| 94 | HPS4 | NA | 11904 | 0.216 | 0.5424 | Yes |
| 95 | EDEM2 | NA | 11981 | 0.215 | 0.5455 | Yes |
| 96 | RUFY3 | NA | 12347 | 0.210 | 0.5433 | Yes |
| 97 | TCAF2 | NA | 12444 | 0.209 | 0.5459 | Yes |
| 98 | TMEM30B | NA | 12811 | 0.203 | 0.5435 | Yes |
| 99 | HRAS | NA | 12965 | 0.201 | 0.5449 | Yes |
| 100 | SLC35D3 | NA | 13131 | 0.199 | 0.5461 | Yes |
| 101 | FBXW7 | NA | 13238 | 0.197 | 0.5483 | Yes |
| 102 | PRKAA1 | NA | 13261 | 0.197 | 0.5521 | Yes |
| 103 | PPP1R13B | NA | 14362 | 0.182 | 0.5357 | No |
| 104 | CASP8 | NA | 14923 | 0.175 | 0.5291 | No |
| 105 | MICALL2 | NA | 15140 | 0.172 | 0.5287 | No |
| 106 | ITGB1BP1 | NA | 15622 | 0.166 | 0.5234 | No |
| 107 | YWHAG | NA | 15800 | 0.164 | 0.5236 | No |
| 108 | CACNB3 | NA | 15801 | 0.164 | 0.5271 | No |
| 109 | RHOU | NA | 16406 | 0.156 | 0.5193 | No |
| 110 | CTDSPL2 | NA | 16435 | 0.156 | 0.5221 | No |
| 111 | CDH1 | NA | 16707 | 0.153 | 0.5203 | No |
| 112 | SMAD3 | NA | 16754 | 0.152 | 0.5227 | No |
| 113 | ZFAND1 | NA | 16811 | 0.151 | 0.5248 | No |
| 114 | CEP131 | NA | 17594 | 0.142 | 0.5135 | No |
| 115 | LEPROT | NA | 17839 | 0.139 | 0.5119 | No |
| 116 | TP53 | NA | 18821 | 0.126 | 0.4965 | No |
| 117 | PRKACA | NA | 19005 | 0.124 | 0.4958 | No |
| 118 | ZPR1 | NA | 19524 | 0.117 | 0.4887 | No |
| 119 | SAE1 | NA | 19684 | 0.115 | 0.4883 | No |
| 120 | RBM22 | NA | 19795 | 0.114 | 0.4886 | No |
| 121 | TMEM30A | NA | 20038 | 0.111 | 0.4865 | No |
| 122 | TP73 | NA | 21933 | 0.089 | 0.4536 | No |
| 123 | SFN | NA | 22639 | 0.081 | 0.4423 | No |
| 124 | BAD | NA | 22872 | 0.079 | 0.4397 | No |
| 125 | PDCD10 | NA | 22962 | 0.078 | 0.4397 | No |
| 126 | TP53BP2 | NA | 23001 | 0.077 | 0.4407 | No |
| 127 | IL1B | NA | 23517 | 0.072 | 0.4327 | No |
| 128 | CAMK1 | NA | 24032 | 0.066 | 0.4247 | No |
| 129 | VAMP2 | NA | 24560 | 0.061 | 0.4163 | No |
| 130 | MIEF2 | NA | 25453 | 0.053 | 0.4010 | No |
| 131 | YWHAE | NA | 25979 | 0.048 | 0.3924 | No |
| 132 | ITGAM | NA | 26116 | 0.047 | 0.3909 | No |
| 133 | B3GAT3 | NA | 26530 | 0.043 | 0.3842 | No |
| 134 | ITGB2 | NA | 26744 | 0.041 | 0.3811 | No |
| 135 | PDZK1 | NA | 27014 | 0.039 | 0.3770 | No |
| 136 | FLNA | NA | 27028 | 0.039 | 0.3776 | No |
| 137 | PRNP | NA | 27554 | 0.036 | 0.3687 | No |
| 138 | CEMIP | NA | 29103 | 0.026 | 0.3408 | No |
| 139 | RAN | NA | 29471 | 0.023 | 0.3346 | No |
| 140 | SLC51B | NA | 29881 | 0.021 | 0.3275 | No |
| 141 | RAC2 | NA | 32687 | 0.008 | 0.2761 | No |
| 142 | OAZ1 | NA | 32935 | 0.007 | 0.2717 | No |
| 143 | TENM1 | NA | 33526 | 0.004 | 0.2610 | No |
| 144 | HTRA2 | NA | 39504 | -0.005 | 0.1512 | No |
| 145 | HCLS1 | NA | 41473 | -0.029 | 0.1157 | No |
| 146 | FIS1 | NA | 41787 | -0.033 | 0.1106 | No |
| 147 | OAZ2 | NA | 41968 | -0.035 | 0.1081 | No |
| 148 | NUTF2 | NA | 42110 | -0.037 | 0.1062 | No |
| 149 | FYN | NA | 43132 | -0.050 | 0.0885 | No |
| 150 | UBL5 | NA | 43532 | -0.055 | 0.0824 | No |
| 151 | HSPA1L | NA | 43708 | -0.058 | 0.0804 | No |
| 152 | CHP2 | NA | 44459 | -0.069 | 0.0680 | No |
| 153 | RAPGEF3 | NA | 45860 | -0.089 | 0.0442 | No |
| 154 | CIB1 | NA | 46701 | -0.102 | 0.0309 | No |
| 155 | TGFB1 | NA | 46703 | -0.102 | 0.0331 | No |
| 156 | ABLIM3 | NA | 46900 | -0.106 | 0.0317 | No |
| 157 | UBE2D3 | NA | 48539 | -0.135 | 0.0044 | No |
| 158 | ZC3H12A | NA | 48550 | -0.136 | 0.0071 | No |
| 159 | GAS6 | NA | 49780 | -0.161 | -0.0120 | No |
| 160 | HSP90AB1 | NA | 50052 | -0.168 | -0.0135 | No |
| 161 | BBC3 | NA | 50088 | -0.168 | -0.0106 | No |
| 162 | EMD | NA | 50517 | -0.177 | -0.0147 | No |
| 163 | UBL4B | NA | 51450 | -0.201 | -0.0276 | No |
| 164 | ANP32B | NA | 51540 | -0.203 | -0.0249 | No |
| 165 | BAG3 | NA | 51657 | -0.207 | -0.0227 | No |
| 166 | SHH | NA | 52917 | -0.241 | -0.0408 | No |
| 167 | TOMM7 | NA | 53191 | -0.255 | -0.0404 | No |
| 168 | IFNG | NA | 53327 | -0.263 | -0.0373 | No |
| 169 | USP36 | NA | 53390 | -0.267 | -0.0328 | No |
| 170 | YWHAH | NA | 53623 | -0.285 | -0.0311 | No |
| 171 | GZMB | NA | 53679 | -0.290 | -0.0260 | No |
| 172 | HPCA | NA | 53791 | -0.300 | -0.0217 | No |
| 173 | PPP3CC | NA | 54266 | -0.382 | -0.0223 | No |
| 174 | PMAIP1 | NA | 54312 | -0.398 | -0.0148 | No |
| 175 | STOM | NA | 54406 | -0.437 | -0.0073 | No |
| 176 | PLK3 | NA | 54503 | -0.504 | 0.0016 | No |