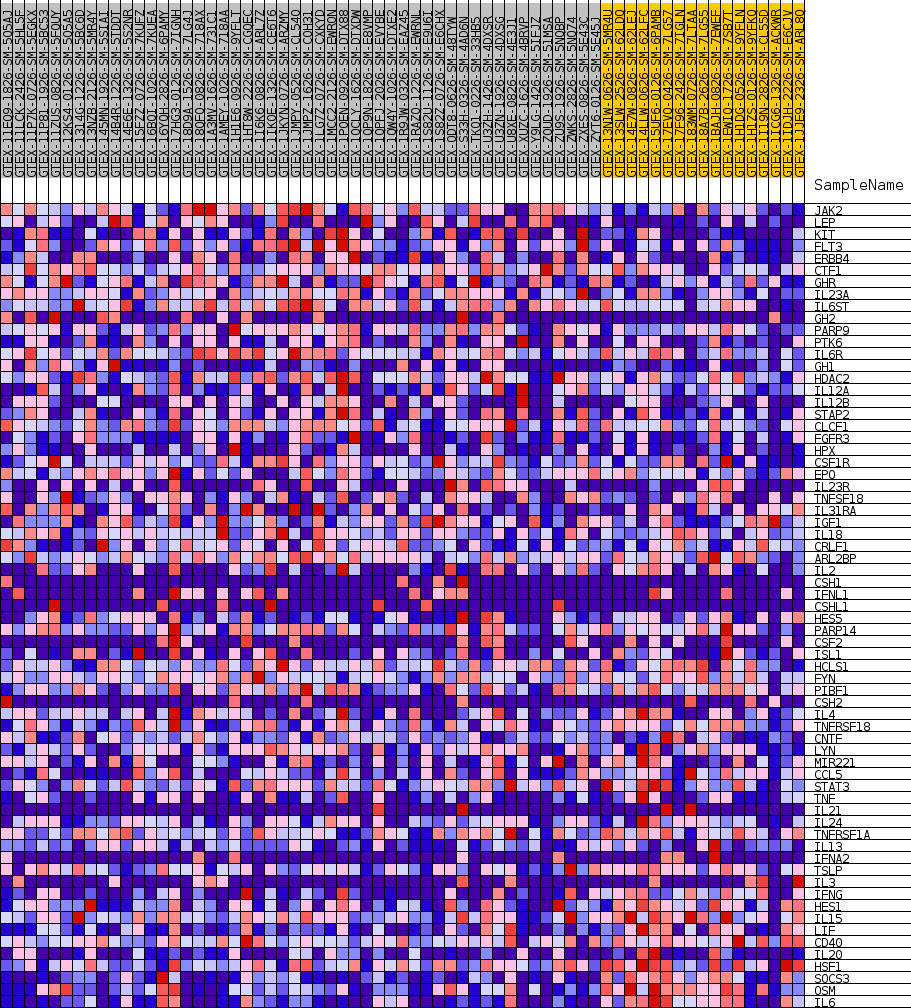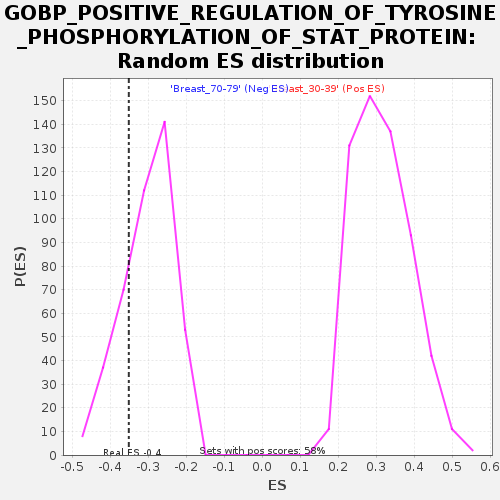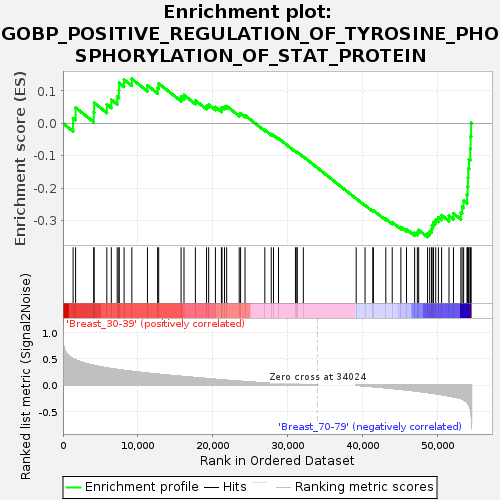
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Breast.Breast.cls #Breast_30-39_versus_Breast_70-79.Breast.cls #Breast_30-39_versus_Breast_70-79_repos |
| Phenotype | Breast.cls#Breast_30-39_versus_Breast_70-79_repos |
| Upregulated in class | Breast_70-79 |
| GeneSet | GOBP_POSITIVE_REGULATION_OF_TYROSINE_PHOSPHORYLATION_OF_STAT_PROTEIN |
| Enrichment Score (ES) | -0.35063064 |
| Normalized Enrichment Score (NES) | -1.1639991 |
| Nominal p-value | 0.2327791 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.999 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | JAK2 | NA | 1350 | 0.499 | 0.0158 | No |
| 2 | LEP | NA | 1684 | 0.477 | 0.0485 | No |
| 3 | KIT | NA | 4102 | 0.371 | 0.0344 | No |
| 4 | FLT3 | NA | 4166 | 0.369 | 0.0632 | No |
| 5 | ERBB4 | NA | 5856 | 0.324 | 0.0586 | No |
| 6 | CTF1 | NA | 6454 | 0.310 | 0.0729 | No |
| 7 | GHR | NA | 7240 | 0.294 | 0.0825 | No |
| 8 | IL23A | NA | 7465 | 0.290 | 0.1019 | No |
| 9 | IL6ST | NA | 7499 | 0.289 | 0.1248 | No |
| 10 | GH2 | NA | 8160 | 0.276 | 0.1352 | No |
| 11 | PARP9 | NA | 9199 | 0.258 | 0.1371 | No |
| 12 | PTK6 | NA | 11281 | 0.225 | 0.1172 | No |
| 13 | IL6R | NA | 12644 | 0.206 | 0.1090 | No |
| 14 | GH1 | NA | 12785 | 0.204 | 0.1230 | No |
| 15 | HDAC2 | NA | 15774 | 0.164 | 0.0816 | No |
| 16 | IL12A | NA | 16167 | 0.159 | 0.0873 | No |
| 17 | IL12B | NA | 17687 | 0.141 | 0.0709 | No |
| 18 | STAP2 | NA | 19180 | 0.121 | 0.0534 | No |
| 19 | CLCF1 | NA | 19463 | 0.118 | 0.0579 | No |
| 20 | FGFR3 | NA | 20365 | 0.107 | 0.0500 | No |
| 21 | HPX | NA | 21168 | 0.098 | 0.0433 | No |
| 22 | CSF1R | NA | 21246 | 0.097 | 0.0497 | No |
| 23 | EPO | NA | 21581 | 0.093 | 0.0511 | No |
| 24 | IL23R | NA | 21868 | 0.090 | 0.0532 | No |
| 25 | TNFSF18 | NA | 23544 | 0.071 | 0.0283 | No |
| 26 | IL31RA | NA | 23728 | 0.070 | 0.0306 | No |
| 27 | IGF1 | NA | 24322 | 0.064 | 0.0249 | No |
| 28 | IL18 | NA | 26968 | 0.040 | -0.0204 | No |
| 29 | CRLF1 | NA | 27826 | 0.034 | -0.0334 | No |
| 30 | ARL2BP | NA | 28125 | 0.032 | -0.0362 | No |
| 31 | IL2 | NA | 28793 | 0.028 | -0.0462 | No |
| 32 | CSH1 | NA | 31060 | 0.015 | -0.0866 | No |
| 33 | IFNL1 | NA | 31251 | 0.014 | -0.0889 | No |
| 34 | CSHL1 | NA | 31292 | 0.014 | -0.0885 | No |
| 35 | HES5 | NA | 32111 | 0.010 | -0.1027 | No |
| 36 | PARP14 | NA | 39174 | -0.001 | -0.2321 | No |
| 37 | CSF2 | NA | 40356 | -0.016 | -0.2525 | No |
| 38 | ISL1 | NA | 41392 | -0.028 | -0.2691 | No |
| 39 | HCLS1 | NA | 41473 | -0.029 | -0.2682 | No |
| 40 | FYN | NA | 43132 | -0.050 | -0.2946 | No |
| 41 | PIBF1 | NA | 43989 | -0.062 | -0.3052 | No |
| 42 | CSH2 | NA | 45145 | -0.078 | -0.3200 | No |
| 43 | IL4 | NA | 45895 | -0.090 | -0.3265 | No |
| 44 | TNFRSF18 | NA | 46984 | -0.107 | -0.3377 | No |
| 45 | CNTF | NA | 47346 | -0.113 | -0.3351 | No |
| 46 | LYN | NA | 47525 | -0.117 | -0.3289 | No |
| 47 | MIR221 | NA | 48712 | -0.139 | -0.3393 | Yes |
| 48 | CCL5 | NA | 48995 | -0.145 | -0.3328 | Yes |
| 49 | STAT3 | NA | 49249 | -0.150 | -0.3252 | Yes |
| 50 | TNF | NA | 49309 | -0.151 | -0.3140 | Yes |
| 51 | IL21 | NA | 49506 | -0.155 | -0.3049 | Yes |
| 52 | IL24 | NA | 49801 | -0.162 | -0.2972 | Yes |
| 53 | TNFRSF1A | NA | 50158 | -0.170 | -0.2899 | Yes |
| 54 | IL13 | NA | 50585 | -0.179 | -0.2831 | Yes |
| 55 | IFNA2 | NA | 51574 | -0.204 | -0.2846 | Yes |
| 56 | TSLP | NA | 52173 | -0.220 | -0.2777 | Yes |
| 57 | IL3 | NA | 53160 | -0.253 | -0.2752 | Yes |
| 58 | IFNG | NA | 53327 | -0.263 | -0.2569 | Yes |
| 59 | HES1 | NA | 53528 | -0.277 | -0.2380 | Yes |
| 60 | IL15 | NA | 53980 | -0.321 | -0.2202 | Yes |
| 61 | LIF | NA | 54056 | -0.333 | -0.1944 | Yes |
| 62 | CD40 | NA | 54122 | -0.346 | -0.1675 | Yes |
| 63 | IL20 | NA | 54142 | -0.351 | -0.1392 | Yes |
| 64 | HSF1 | NA | 54246 | -0.375 | -0.1106 | Yes |
| 65 | SOCS3 | NA | 54415 | -0.440 | -0.0779 | Yes |
| 66 | OSM | NA | 54461 | -0.471 | -0.0404 | Yes |
| 67 | IL6 | NA | 54527 | -0.525 | 0.0012 | Yes |