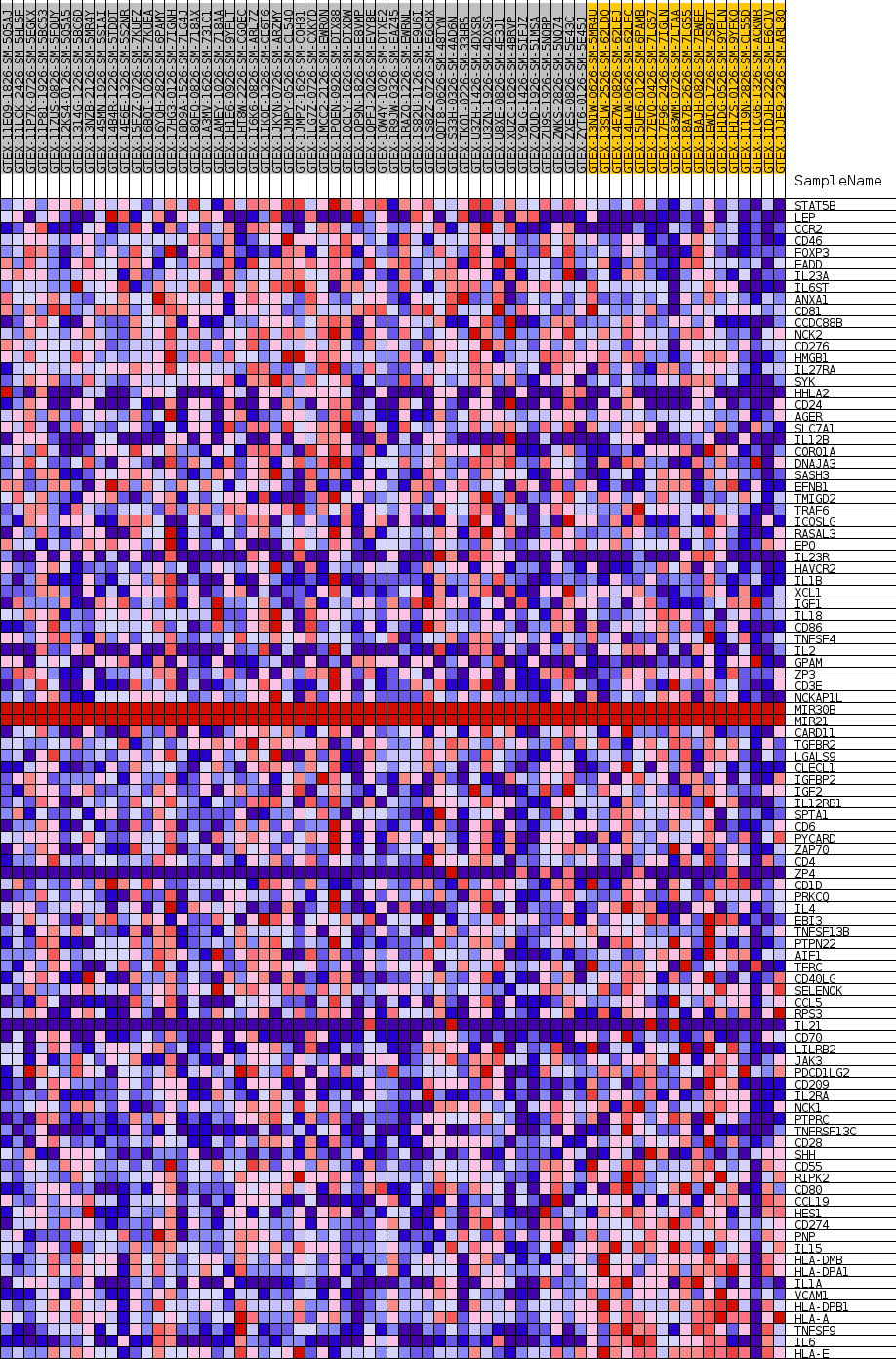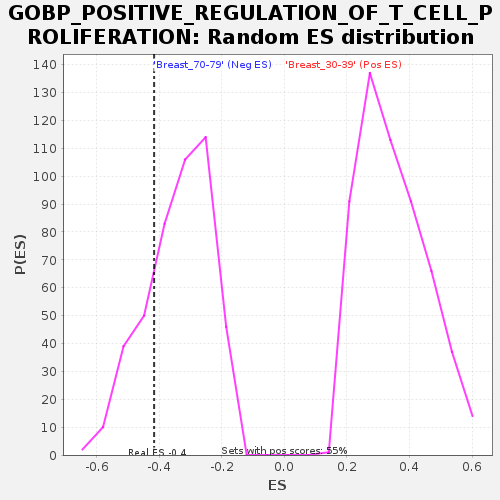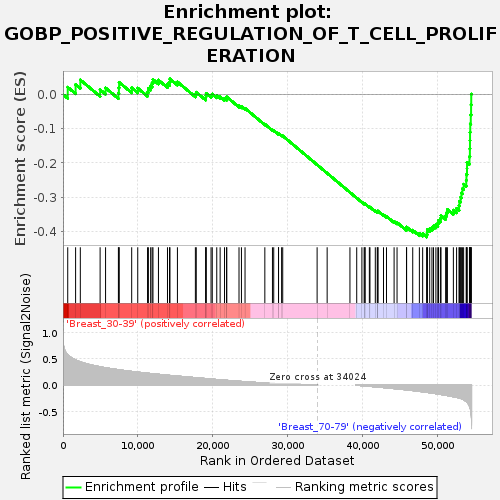
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Breast.Breast.cls #Breast_30-39_versus_Breast_70-79.Breast.cls #Breast_30-39_versus_Breast_70-79_repos |
| Phenotype | Breast.cls#Breast_30-39_versus_Breast_70-79_repos |
| Upregulated in class | Breast_70-79 |
| GeneSet | GOBP_POSITIVE_REGULATION_OF_T_CELL_PROLIFERATION |
| Enrichment Score (ES) | -0.41597036 |
| Normalized Enrichment Score (NES) | -1.2278135 |
| Nominal p-value | 0.22222222 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.998 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | STAT5B | NA | 632 | 0.572 | 0.0210 | No |
| 2 | LEP | NA | 1684 | 0.477 | 0.0289 | No |
| 3 | CCR2 | NA | 2314 | 0.440 | 0.0424 | No |
| 4 | CD46 | NA | 4958 | 0.347 | 0.0136 | No |
| 5 | FOXP3 | NA | 5687 | 0.328 | 0.0190 | No |
| 6 | FADD | NA | 7402 | 0.291 | 0.0041 | No |
| 7 | IL23A | NA | 7465 | 0.290 | 0.0194 | No |
| 8 | IL6ST | NA | 7499 | 0.289 | 0.0353 | No |
| 9 | ANXA1 | NA | 9178 | 0.258 | 0.0192 | No |
| 10 | CD81 | NA | 9987 | 0.245 | 0.0183 | No |
| 11 | CCDC88B | NA | 11296 | 0.225 | 0.0071 | No |
| 12 | NCK2 | NA | 11409 | 0.223 | 0.0178 | No |
| 13 | CD276 | NA | 11712 | 0.218 | 0.0247 | No |
| 14 | HMGB1 | NA | 11903 | 0.216 | 0.0335 | No |
| 15 | IL27RA | NA | 12020 | 0.214 | 0.0436 | No |
| 16 | SYK | NA | 12753 | 0.205 | 0.0418 | No |
| 17 | HHLA2 | NA | 13960 | 0.187 | 0.0303 | No |
| 18 | CD24 | NA | 14231 | 0.183 | 0.0358 | No |
| 19 | AGER | NA | 14292 | 0.183 | 0.0451 | No |
| 20 | SLC7A1 | NA | 15288 | 0.170 | 0.0365 | No |
| 21 | IL12B | NA | 17687 | 0.141 | 0.0006 | No |
| 22 | CORO1A | NA | 17805 | 0.139 | 0.0063 | No |
| 23 | DNAJA3 | NA | 19069 | 0.123 | -0.0098 | No |
| 24 | SASH3 | NA | 19084 | 0.123 | -0.0031 | No |
| 25 | EFNB1 | NA | 19133 | 0.122 | 0.0030 | No |
| 26 | TMIGD2 | NA | 19781 | 0.114 | -0.0024 | No |
| 27 | TRAF6 | NA | 19945 | 0.112 | 0.0010 | No |
| 28 | ICOSLG | NA | 20544 | 0.105 | -0.0040 | No |
| 29 | RASAL3 | NA | 20996 | 0.099 | -0.0066 | No |
| 30 | EPO | NA | 21581 | 0.093 | -0.0121 | No |
| 31 | IL23R | NA | 21868 | 0.090 | -0.0122 | No |
| 32 | HAVCR2 | NA | 21869 | 0.090 | -0.0071 | No |
| 33 | IL1B | NA | 23517 | 0.072 | -0.0333 | No |
| 34 | XCL1 | NA | 23843 | 0.068 | -0.0353 | No |
| 35 | IGF1 | NA | 24322 | 0.064 | -0.0405 | No |
| 36 | IL18 | NA | 26968 | 0.040 | -0.0867 | No |
| 37 | CD86 | NA | 27985 | 0.033 | -0.1035 | No |
| 38 | TNFSF4 | NA | 28156 | 0.032 | -0.1048 | No |
| 39 | IL2 | NA | 28793 | 0.028 | -0.1149 | No |
| 40 | GPAM | NA | 28801 | 0.027 | -0.1135 | No |
| 41 | ZP3 | NA | 29209 | 0.025 | -0.1196 | No |
| 42 | CD3E | NA | 29358 | 0.024 | -0.1209 | No |
| 43 | NCKAP1L | NA | 33953 | 0.001 | -0.2052 | No |
| 44 | MIR30B | NA | 35294 | 0.000 | -0.2297 | No |
| 45 | MIR21 | NA | 38340 | 0.000 | -0.2856 | No |
| 46 | CARD11 | NA | 39245 | -0.002 | -0.3021 | No |
| 47 | TGFBR2 | NA | 39937 | -0.011 | -0.3142 | No |
| 48 | LGALS9 | NA | 40236 | -0.015 | -0.3188 | No |
| 49 | CLECL1 | NA | 40338 | -0.016 | -0.3198 | No |
| 50 | IGFBP2 | NA | 40355 | -0.016 | -0.3192 | No |
| 51 | IGF2 | NA | 40933 | -0.023 | -0.3284 | No |
| 52 | IL12RB1 | NA | 41014 | -0.024 | -0.3285 | No |
| 53 | SPTA1 | NA | 41728 | -0.032 | -0.3398 | No |
| 54 | CD6 | NA | 41988 | -0.035 | -0.3425 | No |
| 55 | PYCARD | NA | 42066 | -0.036 | -0.3419 | No |
| 56 | ZAP70 | NA | 42075 | -0.036 | -0.3400 | No |
| 57 | CD4 | NA | 42823 | -0.046 | -0.3510 | No |
| 58 | ZP4 | NA | 43222 | -0.051 | -0.3554 | No |
| 59 | CD1D | NA | 44239 | -0.065 | -0.3703 | No |
| 60 | PRKCQ | NA | 44633 | -0.071 | -0.3735 | No |
| 61 | IL4 | NA | 45895 | -0.090 | -0.3915 | No |
| 62 | EBI3 | NA | 45913 | -0.090 | -0.3867 | No |
| 63 | TNFSF13B | NA | 46729 | -0.103 | -0.3958 | No |
| 64 | PTPN22 | NA | 47616 | -0.118 | -0.4053 | No |
| 65 | AIF1 | NA | 48055 | -0.127 | -0.4061 | No |
| 66 | TFRC | NA | 48592 | -0.137 | -0.4082 | Yes |
| 67 | CD40LG | NA | 48662 | -0.138 | -0.4016 | Yes |
| 68 | SELENOK | NA | 48675 | -0.138 | -0.3939 | Yes |
| 69 | CCL5 | NA | 48995 | -0.145 | -0.3916 | Yes |
| 70 | RPS3 | NA | 49297 | -0.151 | -0.3885 | Yes |
| 71 | IL21 | NA | 49506 | -0.155 | -0.3834 | Yes |
| 72 | CD70 | NA | 49824 | -0.162 | -0.3800 | Yes |
| 73 | LILRB2 | NA | 50082 | -0.168 | -0.3751 | Yes |
| 74 | JAK3 | NA | 50175 | -0.170 | -0.3671 | Yes |
| 75 | PDCD1LG2 | NA | 50429 | -0.175 | -0.3618 | Yes |
| 76 | CD209 | NA | 50507 | -0.177 | -0.3531 | Yes |
| 77 | IL2RA | NA | 51125 | -0.192 | -0.3535 | Yes |
| 78 | NCK1 | NA | 51252 | -0.196 | -0.3446 | Yes |
| 79 | PTPRC | NA | 51361 | -0.198 | -0.3353 | Yes |
| 80 | TNFRSF13C | NA | 52159 | -0.219 | -0.3375 | Yes |
| 81 | CD28 | NA | 52588 | -0.231 | -0.3321 | Yes |
| 82 | SHH | NA | 52917 | -0.241 | -0.3244 | Yes |
| 83 | CD55 | NA | 52968 | -0.243 | -0.3114 | Yes |
| 84 | RIPK2 | NA | 53126 | -0.251 | -0.3000 | Yes |
| 85 | CD80 | NA | 53227 | -0.256 | -0.2872 | Yes |
| 86 | CCL19 | NA | 53370 | -0.266 | -0.2747 | Yes |
| 87 | HES1 | NA | 53528 | -0.277 | -0.2618 | Yes |
| 88 | CD274 | NA | 53878 | -0.309 | -0.2506 | Yes |
| 89 | PNP | NA | 53905 | -0.312 | -0.2333 | Yes |
| 90 | IL15 | NA | 53980 | -0.321 | -0.2163 | Yes |
| 91 | HLA-DMB | NA | 53985 | -0.322 | -0.1981 | Yes |
| 92 | HLA-DPA1 | NA | 54310 | -0.397 | -0.1814 | Yes |
| 93 | IL1A | NA | 54366 | -0.418 | -0.1586 | Yes |
| 94 | VCAM1 | NA | 54378 | -0.423 | -0.1347 | Yes |
| 95 | HLA-DPB1 | NA | 54388 | -0.429 | -0.1104 | Yes |
| 96 | HLA-A | NA | 54414 | -0.439 | -0.0859 | Yes |
| 97 | TNFSF9 | NA | 54485 | -0.485 | -0.0595 | Yes |
| 98 | IL6 | NA | 54527 | -0.525 | -0.0303 | Yes |
| 99 | HLA-E | NA | 54547 | -0.553 | 0.0008 | Yes |