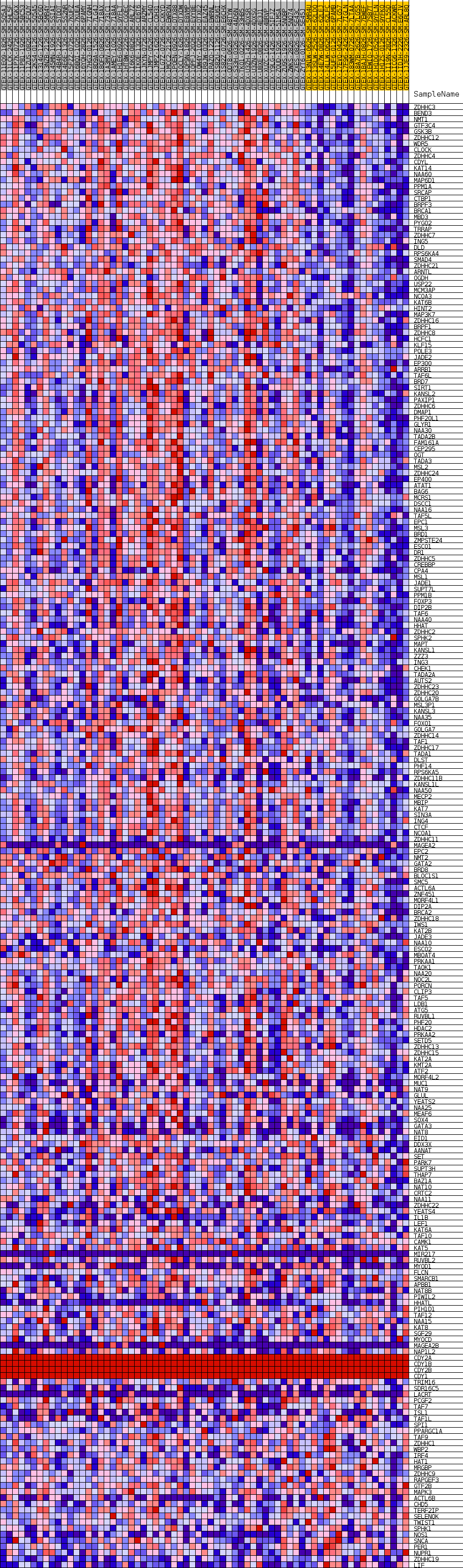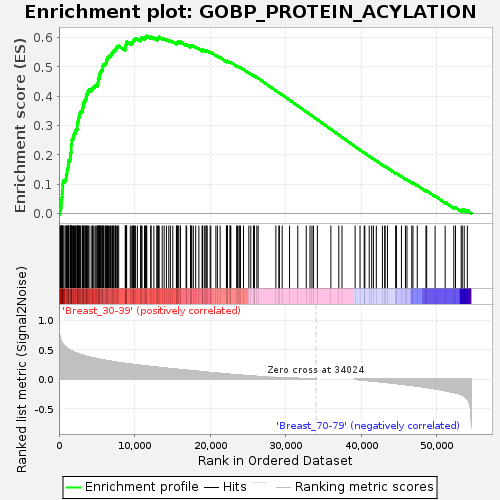
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Breast.Breast.cls #Breast_30-39_versus_Breast_70-79.Breast.cls #Breast_30-39_versus_Breast_70-79_repos |
| Phenotype | Breast.cls#Breast_30-39_versus_Breast_70-79_repos |
| Upregulated in class | Breast_30-39 |
| GeneSet | GOBP_PROTEIN_ACYLATION |
| Enrichment Score (ES) | 0.606111 |
| Normalized Enrichment Score (NES) | 1.6579471 |
| Nominal p-value | 0.005514706 |
| FDR q-value | 0.23616865 |
| FWER p-Value | 0.785 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | ZDHHC3 | NA | 116 | 0.718 | 0.0094 | Yes |
| 2 | BEND3 | NA | 194 | 0.676 | 0.0188 | Yes |
| 3 | NMT1 | NA | 276 | 0.648 | 0.0277 | Yes |
| 4 | GTF3C4 | NA | 320 | 0.632 | 0.0370 | Yes |
| 5 | GSK3B | NA | 325 | 0.632 | 0.0470 | Yes |
| 6 | ZDHHC12 | NA | 385 | 0.617 | 0.0558 | Yes |
| 7 | WDR5 | NA | 412 | 0.611 | 0.0651 | Yes |
| 8 | CLOCK | NA | 429 | 0.608 | 0.0746 | Yes |
| 9 | ZDHHC4 | NA | 478 | 0.598 | 0.0832 | Yes |
| 10 | CDYL | NA | 486 | 0.596 | 0.0927 | Yes |
| 11 | KAT14 | NA | 506 | 0.593 | 0.1018 | Yes |
| 12 | NAA60 | NA | 538 | 0.587 | 0.1107 | Yes |
| 13 | MAP6D1 | NA | 787 | 0.552 | 0.1149 | Yes |
| 14 | PPM1A | NA | 935 | 0.536 | 0.1208 | Yes |
| 15 | SRCAP | NA | 952 | 0.535 | 0.1291 | Yes |
| 16 | CTBP1 | NA | 1036 | 0.527 | 0.1360 | Yes |
| 17 | BRPF3 | NA | 1072 | 0.524 | 0.1437 | Yes |
| 18 | BRCA1 | NA | 1094 | 0.522 | 0.1517 | Yes |
| 19 | MBD3 | NA | 1200 | 0.512 | 0.1580 | Yes |
| 20 | PYGO2 | NA | 1216 | 0.511 | 0.1659 | Yes |
| 21 | TRRAP | NA | 1243 | 0.508 | 0.1736 | Yes |
| 22 | ZDHHC7 | NA | 1258 | 0.507 | 0.1814 | Yes |
| 23 | ING5 | NA | 1450 | 0.492 | 0.1858 | Yes |
| 24 | DLD | NA | 1506 | 0.488 | 0.1926 | Yes |
| 25 | RPS6KA4 | NA | 1516 | 0.487 | 0.2002 | Yes |
| 26 | SMAD4 | NA | 1544 | 0.485 | 0.2075 | Yes |
| 27 | ZDHHC21 | NA | 1609 | 0.482 | 0.2140 | Yes |
| 28 | ARNTL | NA | 1616 | 0.481 | 0.2216 | Yes |
| 29 | OGDH | NA | 1618 | 0.481 | 0.2293 | Yes |
| 30 | USP22 | NA | 1637 | 0.479 | 0.2367 | Yes |
| 31 | MCM3AP | NA | 1696 | 0.476 | 0.2432 | Yes |
| 32 | NCOA3 | NA | 1697 | 0.476 | 0.2508 | Yes |
| 33 | KAT6B | NA | 1808 | 0.467 | 0.2563 | Yes |
| 34 | HINT2 | NA | 1936 | 0.460 | 0.2613 | Yes |
| 35 | MAP3K7 | NA | 1941 | 0.459 | 0.2686 | Yes |
| 36 | ZDHHC16 | NA | 2052 | 0.453 | 0.2738 | Yes |
| 37 | BRPF1 | NA | 2169 | 0.448 | 0.2789 | Yes |
| 38 | ZDHHC8 | NA | 2231 | 0.444 | 0.2849 | Yes |
| 39 | HCFC1 | NA | 2382 | 0.437 | 0.2891 | Yes |
| 40 | KLF15 | NA | 2418 | 0.435 | 0.2954 | Yes |
| 41 | POLE3 | NA | 2420 | 0.434 | 0.3024 | Yes |
| 42 | JADE2 | NA | 2432 | 0.434 | 0.3091 | Yes |
| 43 | EP300 | NA | 2491 | 0.431 | 0.3150 | Yes |
| 44 | ARRB1 | NA | 2571 | 0.428 | 0.3204 | Yes |
| 45 | TAF6L | NA | 2580 | 0.428 | 0.3271 | Yes |
| 46 | BRD7 | NA | 2665 | 0.424 | 0.3323 | Yes |
| 47 | SIRT1 | NA | 2723 | 0.421 | 0.3380 | Yes |
| 48 | KANSL2 | NA | 2787 | 0.418 | 0.3435 | Yes |
| 49 | PAXIP1 | NA | 2868 | 0.414 | 0.3487 | Yes |
| 50 | ZDHHC6 | NA | 3072 | 0.406 | 0.3515 | Yes |
| 51 | DMAP1 | NA | 3110 | 0.405 | 0.3573 | Yes |
| 52 | PHF20L1 | NA | 3169 | 0.403 | 0.3627 | Yes |
| 53 | GLYR1 | NA | 3198 | 0.402 | 0.3686 | Yes |
| 54 | NAA30 | NA | 3229 | 0.401 | 0.3745 | Yes |
| 55 | TADA2B | NA | 3271 | 0.399 | 0.3801 | Yes |
| 56 | FAM161A | NA | 3424 | 0.394 | 0.3836 | Yes |
| 57 | CEP295 | NA | 3464 | 0.392 | 0.3892 | Yes |
| 58 | OGT | NA | 3581 | 0.388 | 0.3933 | Yes |
| 59 | TADA3 | NA | 3592 | 0.388 | 0.3993 | Yes |
| 60 | MSL2 | NA | 3646 | 0.386 | 0.4045 | Yes |
| 61 | ZDHHC24 | NA | 3715 | 0.383 | 0.4094 | Yes |
| 62 | EP400 | NA | 3801 | 0.380 | 0.4139 | Yes |
| 63 | ATAT1 | NA | 3862 | 0.378 | 0.4189 | Yes |
| 64 | BAG6 | NA | 3979 | 0.375 | 0.4227 | Yes |
| 65 | MCRS1 | NA | 4258 | 0.366 | 0.4235 | Yes |
| 66 | DSCC1 | NA | 4421 | 0.362 | 0.4263 | Yes |
| 67 | NAA16 | NA | 4498 | 0.360 | 0.4307 | Yes |
| 68 | TAF5L | NA | 4625 | 0.356 | 0.4340 | Yes |
| 69 | EPC1 | NA | 4803 | 0.351 | 0.4364 | Yes |
| 70 | MSL3 | NA | 4993 | 0.346 | 0.4384 | Yes |
| 71 | BRD1 | NA | 5157 | 0.341 | 0.4409 | Yes |
| 72 | ZMPSTE24 | NA | 5159 | 0.341 | 0.4464 | Yes |
| 73 | ESCO1 | NA | 5184 | 0.340 | 0.4514 | Yes |
| 74 | DR1 | NA | 5189 | 0.340 | 0.4567 | Yes |
| 75 | ZDHHC5 | NA | 5224 | 0.339 | 0.4616 | Yes |
| 76 | CREBBP | NA | 5358 | 0.335 | 0.4645 | Yes |
| 77 | CPA4 | NA | 5390 | 0.335 | 0.4693 | Yes |
| 78 | MSL1 | NA | 5409 | 0.334 | 0.4743 | Yes |
| 79 | JADE1 | NA | 5419 | 0.334 | 0.4795 | Yes |
| 80 | SUPT7L | NA | 5517 | 0.332 | 0.4830 | Yes |
| 81 | PPM1B | NA | 5573 | 0.331 | 0.4873 | Yes |
| 82 | FOXP3 | NA | 5687 | 0.328 | 0.4905 | Yes |
| 83 | DIP2B | NA | 5792 | 0.325 | 0.4938 | Yes |
| 84 | TAF6 | NA | 5802 | 0.325 | 0.4988 | Yes |
| 85 | NAA40 | NA | 5804 | 0.325 | 0.5040 | Yes |
| 86 | HHAT | NA | 5870 | 0.323 | 0.5080 | Yes |
| 87 | ZDHHC2 | NA | 6079 | 0.319 | 0.5092 | Yes |
| 88 | SPHK2 | NA | 6175 | 0.317 | 0.5126 | Yes |
| 89 | MAPT | NA | 6249 | 0.315 | 0.5163 | Yes |
| 90 | KANSL1 | NA | 6295 | 0.314 | 0.5205 | Yes |
| 91 | ZZZ3 | NA | 6298 | 0.314 | 0.5255 | Yes |
| 92 | ING3 | NA | 6421 | 0.311 | 0.5282 | Yes |
| 93 | CHEK1 | NA | 6440 | 0.310 | 0.5328 | Yes |
| 94 | TADA2A | NA | 6552 | 0.308 | 0.5357 | Yes |
| 95 | AUTS2 | NA | 6705 | 0.305 | 0.5378 | Yes |
| 96 | ZDHHC23 | NA | 6827 | 0.303 | 0.5404 | Yes |
| 97 | ZDHHC20 | NA | 6894 | 0.301 | 0.5440 | Yes |
| 98 | GOLGA7B | NA | 7041 | 0.298 | 0.5461 | Yes |
| 99 | MSL3P1 | NA | 7073 | 0.297 | 0.5503 | Yes |
| 100 | KANSL3 | NA | 7177 | 0.296 | 0.5532 | Yes |
| 101 | NAA35 | NA | 7255 | 0.294 | 0.5565 | Yes |
| 102 | FOXO1 | NA | 7408 | 0.291 | 0.5583 | Yes |
| 103 | GOLGA7 | NA | 7487 | 0.289 | 0.5615 | Yes |
| 104 | ZDHHC14 | NA | 7552 | 0.288 | 0.5650 | Yes |
| 105 | TAF1 | NA | 7694 | 0.285 | 0.5669 | Yes |
| 106 | ZDHHC17 | NA | 7732 | 0.284 | 0.5708 | Yes |
| 107 | TADA1 | NA | 7901 | 0.281 | 0.5722 | Yes |
| 108 | DLST | NA | 8773 | 0.265 | 0.5604 | Yes |
| 109 | PHF14 | NA | 8798 | 0.265 | 0.5642 | Yes |
| 110 | RPS6KA5 | NA | 8811 | 0.265 | 0.5683 | Yes |
| 111 | ZDHHC11B | NA | 8815 | 0.265 | 0.5724 | Yes |
| 112 | KANSL1L | NA | 8834 | 0.264 | 0.5763 | Yes |
| 113 | NAA50 | NA | 8911 | 0.263 | 0.5791 | Yes |
| 114 | MECP2 | NA | 8932 | 0.262 | 0.5830 | Yes |
| 115 | MBIP | NA | 8951 | 0.262 | 0.5868 | Yes |
| 116 | KAT7 | NA | 9435 | 0.254 | 0.5820 | Yes |
| 117 | SIN3A | NA | 9645 | 0.250 | 0.5822 | Yes |
| 118 | ING4 | NA | 9698 | 0.249 | 0.5852 | Yes |
| 119 | CTCF | NA | 9804 | 0.248 | 0.5873 | Yes |
| 120 | NCOA1 | NA | 9873 | 0.247 | 0.5900 | Yes |
| 121 | ZDHHC11 | NA | 9920 | 0.246 | 0.5930 | Yes |
| 122 | MAGEA2 | NA | 10020 | 0.244 | 0.5951 | Yes |
| 123 | EPC2 | NA | 10139 | 0.242 | 0.5968 | Yes |
| 124 | NMT2 | NA | 10381 | 0.238 | 0.5962 | Yes |
| 125 | GATA2 | NA | 10795 | 0.232 | 0.5923 | Yes |
| 126 | BRD8 | NA | 10844 | 0.231 | 0.5952 | Yes |
| 127 | BLOC1S1 | NA | 10862 | 0.231 | 0.5985 | Yes |
| 128 | SMC5 | NA | 10993 | 0.229 | 0.5998 | Yes |
| 129 | ACTL6A | NA | 11317 | 0.224 | 0.5975 | Yes |
| 130 | ZNF451 | NA | 11418 | 0.223 | 0.5992 | Yes |
| 131 | MORF4L1 | NA | 11468 | 0.222 | 0.6019 | Yes |
| 132 | DIP2A | NA | 11573 | 0.221 | 0.6035 | Yes |
| 133 | BRCA2 | NA | 11622 | 0.220 | 0.6061 | Yes |
| 134 | ZDHHC18 | NA | 12142 | 0.213 | 0.6000 | No |
| 135 | IWS1 | NA | 12187 | 0.212 | 0.6026 | No |
| 136 | KAT2B | NA | 12557 | 0.207 | 0.5991 | No |
| 137 | JADE3 | NA | 12970 | 0.201 | 0.5947 | No |
| 138 | NAA10 | NA | 12977 | 0.201 | 0.5978 | No |
| 139 | ESCO2 | NA | 13102 | 0.199 | 0.5987 | No |
| 140 | MBOAT4 | NA | 13156 | 0.199 | 0.6010 | No |
| 141 | PRKAA1 | NA | 13261 | 0.197 | 0.6022 | No |
| 142 | TAOK1 | NA | 13663 | 0.191 | 0.5979 | No |
| 143 | NAA20 | NA | 13922 | 0.188 | 0.5961 | No |
| 144 | NOC2L | NA | 14222 | 0.184 | 0.5936 | No |
| 145 | PORCN | NA | 14535 | 0.180 | 0.5907 | No |
| 146 | CLIP3 | NA | 14738 | 0.177 | 0.5898 | No |
| 147 | TAF5 | NA | 15065 | 0.173 | 0.5866 | No |
| 148 | LDB1 | NA | 15544 | 0.167 | 0.5805 | No |
| 149 | ATG5 | NA | 15552 | 0.167 | 0.5830 | No |
| 150 | RUVBL1 | NA | 15713 | 0.165 | 0.5827 | No |
| 151 | PHF20 | NA | 15745 | 0.165 | 0.5848 | No |
| 152 | HDAC2 | NA | 15774 | 0.164 | 0.5869 | No |
| 153 | PRKAA2 | NA | 16056 | 0.161 | 0.5843 | No |
| 154 | SETD5 | NA | 16066 | 0.161 | 0.5867 | No |
| 155 | ZDHHC13 | NA | 16823 | 0.151 | 0.5752 | No |
| 156 | ZDHHC15 | NA | 16900 | 0.150 | 0.5762 | No |
| 157 | KAT2A | NA | 17405 | 0.144 | 0.5693 | No |
| 158 | KMT2A | NA | 17412 | 0.144 | 0.5715 | No |
| 159 | ATF2 | NA | 17487 | 0.143 | 0.5724 | No |
| 160 | MORF4L2 | NA | 17570 | 0.142 | 0.5732 | No |
| 161 | MUC1 | NA | 17782 | 0.139 | 0.5715 | No |
| 162 | NAT9 | NA | 18096 | 0.135 | 0.5679 | No |
| 163 | GLUL | NA | 18517 | 0.130 | 0.5623 | No |
| 164 | YEATS2 | NA | 18943 | 0.124 | 0.5565 | No |
| 165 | NAA25 | NA | 18999 | 0.124 | 0.5574 | No |
| 166 | MEAF6 | NA | 19027 | 0.123 | 0.5589 | No |
| 167 | SOX4 | NA | 19301 | 0.120 | 0.5558 | No |
| 168 | GATA3 | NA | 19337 | 0.120 | 0.5571 | No |
| 169 | NAT8 | NA | 19560 | 0.117 | 0.5549 | No |
| 170 | EID1 | NA | 19609 | 0.116 | 0.5559 | No |
| 171 | DDX3X | NA | 20006 | 0.111 | 0.5504 | No |
| 172 | AANAT | NA | 20099 | 0.110 | 0.5504 | No |
| 173 | SET | NA | 20765 | 0.102 | 0.5398 | No |
| 174 | PARK7 | NA | 20966 | 0.100 | 0.5377 | No |
| 175 | SUPT3H | NA | 21329 | 0.096 | 0.5326 | No |
| 176 | THAP7 | NA | 22162 | 0.087 | 0.5187 | No |
| 177 | BAZ1A | NA | 22202 | 0.086 | 0.5194 | No |
| 178 | NAT10 | NA | 22266 | 0.085 | 0.5196 | No |
| 179 | CRTC2 | NA | 22289 | 0.085 | 0.5205 | No |
| 180 | NAA11 | NA | 22631 | 0.081 | 0.5156 | No |
| 181 | ZDHHC22 | NA | 22678 | 0.081 | 0.5160 | No |
| 182 | YEATS4 | NA | 22748 | 0.080 | 0.5160 | No |
| 183 | IL1B | NA | 23517 | 0.072 | 0.5030 | No |
| 184 | LEF1 | NA | 23620 | 0.071 | 0.5023 | No |
| 185 | KAT6A | NA | 23822 | 0.069 | 0.4997 | No |
| 186 | TAF10 | NA | 23933 | 0.068 | 0.4987 | No |
| 187 | CAMK1 | NA | 24032 | 0.066 | 0.4980 | No |
| 188 | KAT5 | NA | 24425 | 0.063 | 0.4918 | No |
| 189 | MIR217 | NA | 25131 | 0.056 | 0.4797 | No |
| 190 | RUVBL2 | NA | 25390 | 0.053 | 0.4758 | No |
| 191 | MYOD1 | NA | 25747 | 0.050 | 0.4701 | No |
| 192 | FLCN | NA | 25822 | 0.049 | 0.4695 | No |
| 193 | SMARCB1 | NA | 25864 | 0.049 | 0.4695 | No |
| 194 | APBB1 | NA | 26192 | 0.046 | 0.4643 | No |
| 195 | NAT8B | NA | 26363 | 0.045 | 0.4619 | No |
| 196 | PIWIL2 | NA | 28706 | 0.028 | 0.4192 | No |
| 197 | HHATL | NA | 29108 | 0.026 | 0.4122 | No |
| 198 | PIH1D1 | NA | 29171 | 0.025 | 0.4115 | No |
| 199 | TAF12 | NA | 29544 | 0.023 | 0.4050 | No |
| 200 | NAA15 | NA | 30519 | 0.018 | 0.3874 | No |
| 201 | KAT8 | NA | 31609 | 0.013 | 0.3676 | No |
| 202 | SGF29 | NA | 32733 | 0.008 | 0.3470 | No |
| 203 | MYOCD | NA | 33228 | 0.006 | 0.3380 | No |
| 204 | MAGEA2B | NA | 33487 | 0.005 | 0.3333 | No |
| 205 | NAP1L2 | NA | 33680 | 0.003 | 0.3299 | No |
| 206 | CDY2A | NA | 34203 | 0.000 | 0.3203 | No |
| 207 | CDY1B | NA | 35991 | 0.000 | 0.2874 | No |
| 208 | CDY2B | NA | 37040 | 0.000 | 0.2681 | No |
| 209 | CDY1 | NA | 37465 | 0.000 | 0.2603 | No |
| 210 | TRIM16 | NA | 39199 | -0.001 | 0.2284 | No |
| 211 | SDR16C5 | NA | 39851 | -0.010 | 0.2166 | No |
| 212 | LACRT | NA | 40405 | -0.017 | 0.2067 | No |
| 213 | PCGF2 | NA | 40487 | -0.018 | 0.2055 | No |
| 214 | TAF7 | NA | 41067 | -0.025 | 0.1952 | No |
| 215 | ISL1 | NA | 41392 | -0.028 | 0.1897 | No |
| 216 | TAF1L | NA | 41622 | -0.031 | 0.1860 | No |
| 217 | SPI1 | NA | 42005 | -0.035 | 0.1796 | No |
| 218 | PPARGC1A | NA | 42813 | -0.046 | 0.1654 | No |
| 219 | TAF9 | NA | 43119 | -0.050 | 0.1606 | No |
| 220 | ZDHHC1 | NA | 43172 | -0.051 | 0.1605 | No |
| 221 | WBP2 | NA | 43483 | -0.055 | 0.1557 | No |
| 222 | IRF4 | NA | 44587 | -0.070 | 0.1365 | No |
| 223 | HAT1 | NA | 44618 | -0.071 | 0.1371 | No |
| 224 | MRGBP | NA | 44658 | -0.071 | 0.1375 | No |
| 225 | ZDHHC9 | NA | 45333 | -0.081 | 0.1264 | No |
| 226 | RAPGEF3 | NA | 45860 | -0.089 | 0.1182 | No |
| 227 | GTF2B | NA | 46063 | -0.092 | 0.1159 | No |
| 228 | MAPK3 | NA | 46700 | -0.102 | 0.1059 | No |
| 229 | ACTL6B | NA | 46858 | -0.105 | 0.1046 | No |
| 230 | CHD5 | NA | 47426 | -0.115 | 0.0961 | No |
| 231 | TERF2IP | NA | 48578 | -0.136 | 0.0771 | No |
| 232 | SELENOK | NA | 48675 | -0.138 | 0.0775 | No |
| 233 | TWIST1 | NA | 49789 | -0.162 | 0.0596 | No |
| 234 | SPHK1 | NA | 51119 | -0.192 | 0.0382 | No |
| 235 | NOS1 | NA | 52264 | -0.222 | 0.0208 | No |
| 236 | SNCA | NA | 52490 | -0.228 | 0.0203 | No |
| 237 | PER1 | NA | 53244 | -0.257 | 0.0105 | No |
| 238 | NUPR1 | NA | 53377 | -0.266 | 0.0124 | No |
| 239 | ZDHHC19 | NA | 53642 | -0.286 | 0.0121 | No |
| 240 | LIF | NA | 54056 | -0.333 | 0.0098 | No |