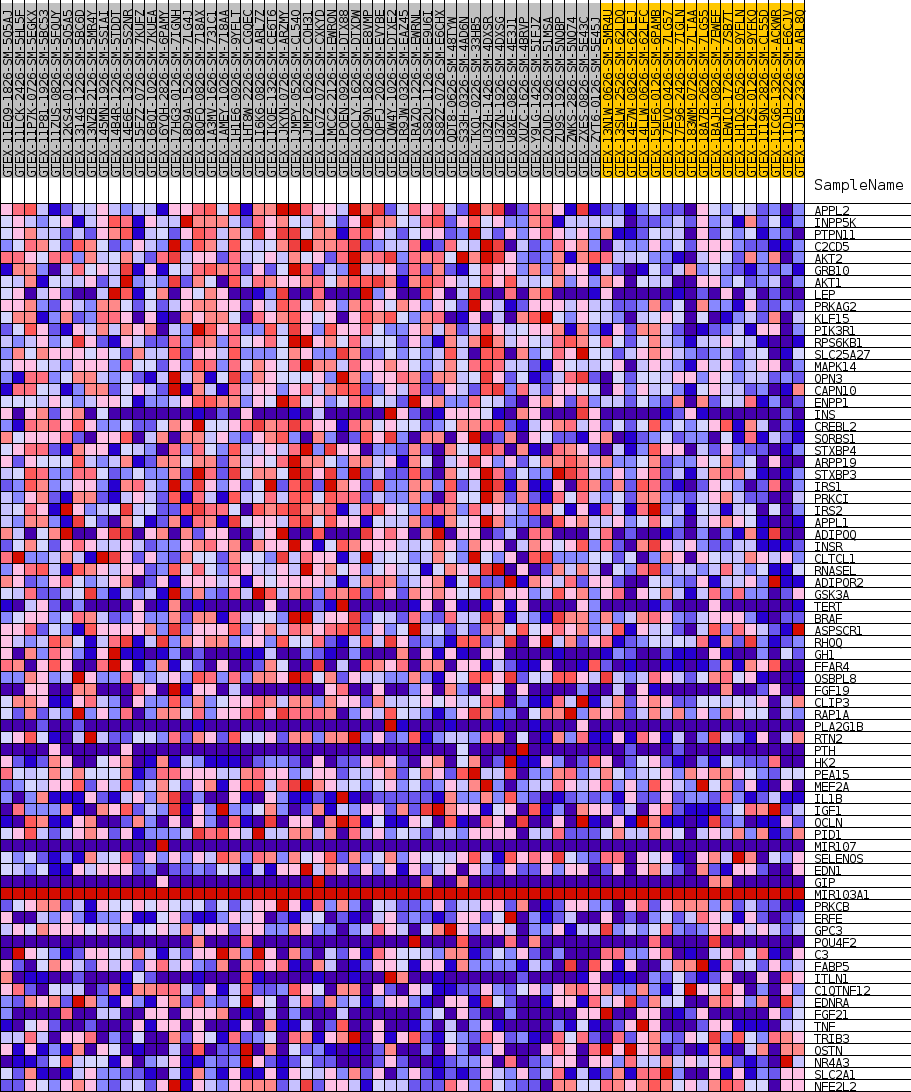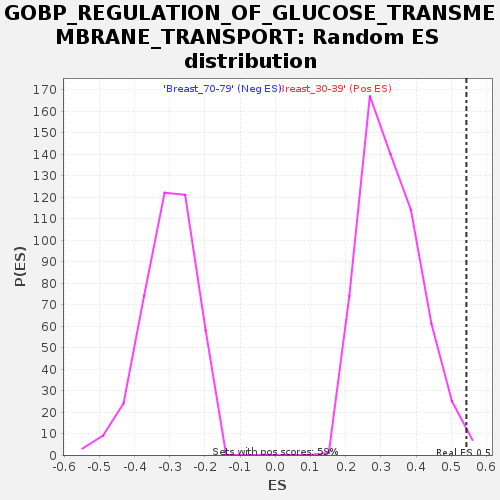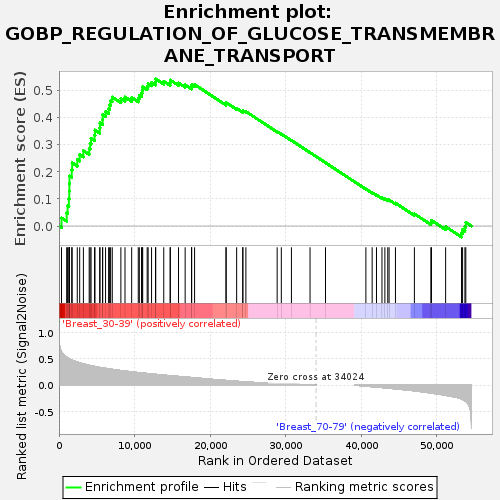
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Breast.Breast.cls #Breast_30-39_versus_Breast_70-79.Breast.cls #Breast_30-39_versus_Breast_70-79_repos |
| Phenotype | Breast.cls#Breast_30-39_versus_Breast_70-79_repos |
| Upregulated in class | Breast_30-39 |
| GeneSet | GOBP_REGULATION_OF_GLUCOSE_TRANSMEMBRANE_TRANSPORT |
| Enrichment Score (ES) | 0.54318255 |
| Normalized Enrichment Score (NES) | 1.6464493 |
| Nominal p-value | 0.0067911716 |
| FDR q-value | 0.24233742 |
| FWER p-Value | 0.812 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | APPL2 | NA | 333 | 0.629 | 0.0300 | Yes |
| 2 | INPP5K | NA | 1017 | 0.528 | 0.0478 | Yes |
| 3 | PTPN11 | NA | 1141 | 0.517 | 0.0752 | Yes |
| 4 | C2CD5 | NA | 1305 | 0.503 | 0.1011 | Yes |
| 5 | AKT2 | NA | 1358 | 0.498 | 0.1287 | Yes |
| 6 | GRB10 | NA | 1373 | 0.497 | 0.1570 | Yes |
| 7 | AKT1 | NA | 1390 | 0.496 | 0.1851 | Yes |
| 8 | LEP | NA | 1684 | 0.477 | 0.2071 | Yes |
| 9 | PRKAG2 | NA | 1741 | 0.473 | 0.2332 | Yes |
| 10 | KLF15 | NA | 2418 | 0.435 | 0.2458 | Yes |
| 11 | PIK3R1 | NA | 2739 | 0.420 | 0.2640 | Yes |
| 12 | RPS6KB1 | NA | 3228 | 0.401 | 0.2781 | Yes |
| 13 | SLC25A27 | NA | 3996 | 0.374 | 0.2855 | Yes |
| 14 | MAPK14 | NA | 4118 | 0.371 | 0.3045 | Yes |
| 15 | OPN3 | NA | 4244 | 0.367 | 0.3233 | Yes |
| 16 | CAPN10 | NA | 4702 | 0.353 | 0.3352 | Yes |
| 17 | ENPP1 | NA | 4763 | 0.352 | 0.3543 | Yes |
| 18 | INS | NA | 5395 | 0.335 | 0.3619 | Yes |
| 19 | CREBL2 | NA | 5424 | 0.334 | 0.3805 | Yes |
| 20 | SORBS1 | NA | 5768 | 0.326 | 0.3929 | Yes |
| 21 | STXBP4 | NA | 5781 | 0.326 | 0.4114 | Yes |
| 22 | ARPP19 | NA | 6164 | 0.317 | 0.4226 | Yes |
| 23 | STXBP3 | NA | 6567 | 0.308 | 0.4329 | Yes |
| 24 | IRS1 | NA | 6700 | 0.305 | 0.4479 | Yes |
| 25 | PRKCI | NA | 6833 | 0.302 | 0.4629 | Yes |
| 26 | IRS2 | NA | 7052 | 0.298 | 0.4760 | Yes |
| 27 | APPL1 | NA | 8192 | 0.275 | 0.4709 | Yes |
| 28 | ADIPOQ | NA | 8737 | 0.266 | 0.4762 | Yes |
| 29 | INSR | NA | 9620 | 0.250 | 0.4744 | Yes |
| 30 | CLTCL1 | NA | 10491 | 0.237 | 0.4720 | Yes |
| 31 | RNASEL | NA | 10634 | 0.234 | 0.4828 | Yes |
| 32 | ADIPOR2 | NA | 10925 | 0.230 | 0.4907 | Yes |
| 33 | GSK3A | NA | 11015 | 0.229 | 0.5022 | Yes |
| 34 | TERT | NA | 11079 | 0.228 | 0.5141 | Yes |
| 35 | BRAF | NA | 11673 | 0.219 | 0.5158 | Yes |
| 36 | ASPSCR1 | NA | 11798 | 0.217 | 0.5260 | Yes |
| 37 | RHOQ | NA | 12252 | 0.211 | 0.5298 | Yes |
| 38 | GH1 | NA | 12785 | 0.204 | 0.5318 | Yes |
| 39 | FFAR4 | NA | 12800 | 0.204 | 0.5432 | Yes |
| 40 | OSBPL8 | NA | 13867 | 0.188 | 0.5344 | No |
| 41 | FGF19 | NA | 14713 | 0.178 | 0.5291 | No |
| 42 | CLIP3 | NA | 14738 | 0.177 | 0.5388 | No |
| 43 | RAP1A | NA | 15821 | 0.164 | 0.5284 | No |
| 44 | PLA2G1B | NA | 16700 | 0.153 | 0.5211 | No |
| 45 | RTN2 | NA | 17542 | 0.143 | 0.5138 | No |
| 46 | PTH | NA | 17567 | 0.142 | 0.5215 | No |
| 47 | HK2 | NA | 17949 | 0.137 | 0.5224 | No |
| 48 | PEA15 | NA | 22105 | 0.087 | 0.4512 | No |
| 49 | MEF2A | NA | 22129 | 0.087 | 0.4558 | No |
| 50 | IL1B | NA | 23517 | 0.072 | 0.4344 | No |
| 51 | IGF1 | NA | 24322 | 0.064 | 0.4234 | No |
| 52 | OCLN | NA | 24346 | 0.064 | 0.4266 | No |
| 53 | PID1 | NA | 24739 | 0.060 | 0.4228 | No |
| 54 | MIR107 | NA | 28878 | 0.027 | 0.3485 | No |
| 55 | SELENOS | NA | 29430 | 0.024 | 0.3397 | No |
| 56 | EDN1 | NA | 30768 | 0.016 | 0.3161 | No |
| 57 | GIP | NA | 33232 | 0.006 | 0.2713 | No |
| 58 | MIR103A1 | NA | 35279 | 0.000 | 0.2337 | No |
| 59 | PRKCB | NA | 40625 | -0.019 | 0.1368 | No |
| 60 | ERFE | NA | 41455 | -0.029 | 0.1233 | No |
| 61 | GPC3 | NA | 42028 | -0.036 | 0.1148 | No |
| 62 | POU4F2 | NA | 42754 | -0.045 | 0.1041 | No |
| 63 | C3 | NA | 43139 | -0.050 | 0.1000 | No |
| 64 | FABP5 | NA | 43523 | -0.055 | 0.0961 | No |
| 65 | ITLN1 | NA | 43696 | -0.058 | 0.0963 | No |
| 66 | C1QTNF12 | NA | 44539 | -0.070 | 0.0848 | No |
| 67 | EDNRA | NA | 47047 | -0.108 | 0.0451 | No |
| 68 | FGF21 | NA | 49225 | -0.149 | 0.0137 | No |
| 69 | TNF | NA | 49309 | -0.151 | 0.0208 | No |
| 70 | TRIB3 | NA | 51182 | -0.194 | -0.0024 | No |
| 71 | OSTN | NA | 53286 | -0.260 | -0.0260 | No |
| 72 | NR4A3 | NA | 53426 | -0.270 | -0.0131 | No |
| 73 | SLC2A1 | NA | 53736 | -0.294 | -0.0019 | No |
| 74 | NFE2L2 | NA | 53852 | -0.306 | 0.0136 | No |