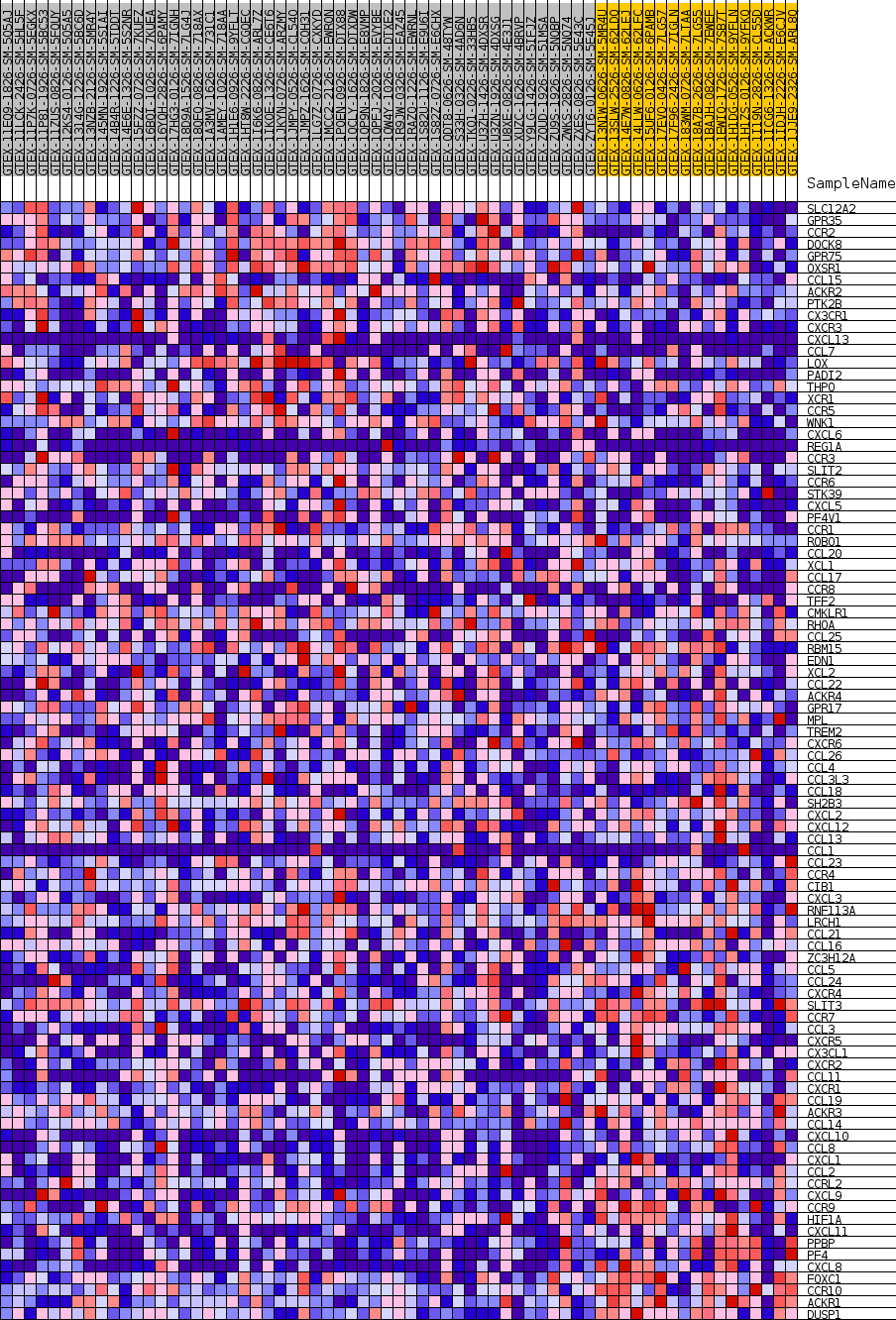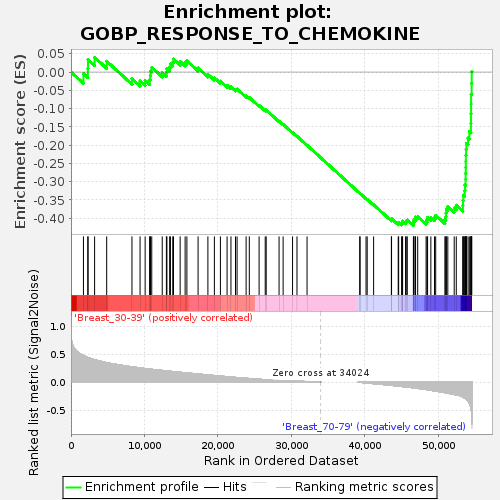
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Breast.Breast.cls #Breast_30-39_versus_Breast_70-79.Breast.cls #Breast_30-39_versus_Breast_70-79_repos |
| Phenotype | Breast.cls#Breast_30-39_versus_Breast_70-79_repos |
| Upregulated in class | Breast_70-79 |
| GeneSet | GOBP_RESPONSE_TO_CHEMOKINE |
| Enrichment Score (ES) | -0.42017996 |
| Normalized Enrichment Score (NES) | -1.3334147 |
| Nominal p-value | 0.109725684 |
| FDR q-value | 0.99405473 |
| FWER p-Value | 0.997 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | SLC12A2 | NA | 1694 | 0.476 | -0.0043 | No |
| 2 | GPR35 | NA | 2299 | 0.440 | 0.0094 | No |
| 3 | CCR2 | NA | 2314 | 0.440 | 0.0339 | No |
| 4 | DOCK8 | NA | 3214 | 0.402 | 0.0400 | No |
| 5 | GPR75 | NA | 4858 | 0.349 | 0.0295 | No |
| 6 | OXSR1 | NA | 8299 | 0.274 | -0.0182 | No |
| 7 | CCL15 | NA | 9403 | 0.254 | -0.0241 | No |
| 8 | ACKR2 | NA | 10095 | 0.243 | -0.0231 | No |
| 9 | PTK2B | NA | 10700 | 0.234 | -0.0211 | No |
| 10 | CX3CR1 | NA | 10796 | 0.232 | -0.0098 | No |
| 11 | CXCR3 | NA | 10833 | 0.231 | 0.0026 | No |
| 12 | CXCL13 | NA | 10995 | 0.229 | 0.0125 | No |
| 13 | CCL7 | NA | 12422 | 0.209 | -0.0019 | No |
| 14 | LOX | NA | 12989 | 0.201 | -0.0009 | No |
| 15 | PADI2 | NA | 13057 | 0.200 | 0.0091 | No |
| 16 | THPO | NA | 13432 | 0.195 | 0.0132 | No |
| 17 | XCR1 | NA | 13539 | 0.193 | 0.0221 | No |
| 18 | CCR5 | NA | 13857 | 0.189 | 0.0269 | No |
| 19 | WNK1 | NA | 13954 | 0.187 | 0.0357 | No |
| 20 | CXCL6 | NA | 14860 | 0.176 | 0.0289 | No |
| 21 | REG1A | NA | 15539 | 0.167 | 0.0259 | No |
| 22 | CCR3 | NA | 15779 | 0.164 | 0.0308 | No |
| 23 | SLIT2 | NA | 17302 | 0.145 | 0.0110 | No |
| 24 | CCR6 | NA | 18639 | 0.128 | -0.0063 | No |
| 25 | STK39 | NA | 19522 | 0.117 | -0.0159 | No |
| 26 | CXCL5 | NA | 20343 | 0.107 | -0.0249 | No |
| 27 | PF4V1 | NA | 21251 | 0.097 | -0.0361 | No |
| 28 | CCR1 | NA | 21778 | 0.090 | -0.0406 | No |
| 29 | ROBO1 | NA | 22398 | 0.084 | -0.0473 | No |
| 30 | CCL20 | NA | 22612 | 0.081 | -0.0466 | No |
| 31 | XCL1 | NA | 23843 | 0.068 | -0.0653 | No |
| 32 | CCL17 | NA | 24278 | 0.064 | -0.0697 | No |
| 33 | CCR8 | NA | 25610 | 0.051 | -0.0912 | No |
| 34 | TFF2 | NA | 26443 | 0.044 | -0.1040 | No |
| 35 | CMKLR1 | NA | 26577 | 0.043 | -0.1040 | No |
| 36 | RHOA | NA | 28328 | 0.030 | -0.1344 | No |
| 37 | CCL25 | NA | 28881 | 0.027 | -0.1430 | No |
| 38 | RBM15 | NA | 30165 | 0.019 | -0.1655 | No |
| 39 | EDN1 | NA | 30768 | 0.016 | -0.1756 | No |
| 40 | XCL2 | NA | 32139 | 0.010 | -0.2002 | No |
| 41 | CCL22 | NA | 39310 | -0.003 | -0.3316 | No |
| 42 | ACKR4 | NA | 39361 | -0.003 | -0.3323 | No |
| 43 | GPR17 | NA | 40170 | -0.014 | -0.3464 | No |
| 44 | MPL | NA | 40326 | -0.016 | -0.3484 | No |
| 45 | TREM2 | NA | 41202 | -0.026 | -0.3629 | No |
| 46 | CXCR6 | NA | 43620 | -0.057 | -0.4041 | No |
| 47 | CCL26 | NA | 43644 | -0.057 | -0.4013 | No |
| 48 | CCL4 | NA | 44550 | -0.070 | -0.4140 | No |
| 49 | CCL3L3 | NA | 44612 | -0.071 | -0.4111 | No |
| 50 | CCL18 | NA | 45013 | -0.077 | -0.4142 | No |
| 51 | SH2B3 | NA | 45107 | -0.078 | -0.4115 | No |
| 52 | CXCL2 | NA | 45149 | -0.078 | -0.4078 | No |
| 53 | CXCL12 | NA | 45552 | -0.084 | -0.4105 | No |
| 54 | CCL13 | NA | 45642 | -0.086 | -0.4073 | No |
| 55 | CCL1 | NA | 45784 | -0.088 | -0.4049 | No |
| 56 | CCL23 | NA | 46617 | -0.101 | -0.4145 | Yes |
| 57 | CCR4 | NA | 46637 | -0.101 | -0.4092 | Yes |
| 58 | CIB1 | NA | 46701 | -0.102 | -0.4046 | Yes |
| 59 | CXCL3 | NA | 46874 | -0.105 | -0.4018 | Yes |
| 60 | RNF113A | NA | 46912 | -0.106 | -0.3965 | Yes |
| 61 | LRCH1 | NA | 47204 | -0.111 | -0.3956 | Yes |
| 62 | CCL21 | NA | 48347 | -0.132 | -0.4091 | Yes |
| 63 | CCL16 | NA | 48475 | -0.135 | -0.4039 | Yes |
| 64 | ZC3H12A | NA | 48550 | -0.136 | -0.3976 | Yes |
| 65 | CCL5 | NA | 48995 | -0.145 | -0.3976 | Yes |
| 66 | CCL24 | NA | 49494 | -0.155 | -0.3980 | Yes |
| 67 | CXCR4 | NA | 49654 | -0.158 | -0.3920 | Yes |
| 68 | SLIT3 | NA | 50908 | -0.187 | -0.4045 | Yes |
| 69 | CCR7 | NA | 51037 | -0.190 | -0.3961 | Yes |
| 70 | CCL3 | NA | 51058 | -0.191 | -0.3858 | Yes |
| 71 | CXCR5 | NA | 51093 | -0.192 | -0.3756 | Yes |
| 72 | CX3CL1 | NA | 51304 | -0.197 | -0.3684 | Yes |
| 73 | CXCR2 | NA | 52174 | -0.220 | -0.3719 | Yes |
| 74 | CCL11 | NA | 52457 | -0.227 | -0.3643 | Yes |
| 75 | CXCR1 | NA | 53355 | -0.265 | -0.3659 | Yes |
| 76 | CCL19 | NA | 53370 | -0.266 | -0.3512 | Yes |
| 77 | ACKR3 | NA | 53419 | -0.269 | -0.3369 | Yes |
| 78 | CCL14 | NA | 53600 | -0.283 | -0.3243 | Yes |
| 79 | CXCL10 | NA | 53640 | -0.286 | -0.3089 | Yes |
| 80 | CCL8 | NA | 53728 | -0.293 | -0.2940 | Yes |
| 81 | CXCL1 | NA | 53731 | -0.294 | -0.2775 | Yes |
| 82 | CCL2 | NA | 53749 | -0.296 | -0.2611 | Yes |
| 83 | CCRL2 | NA | 53755 | -0.297 | -0.2445 | Yes |
| 84 | CXCL9 | NA | 53759 | -0.297 | -0.2278 | Yes |
| 85 | CCR9 | NA | 53794 | -0.301 | -0.2115 | Yes |
| 86 | HIF1A | NA | 53833 | -0.305 | -0.1950 | Yes |
| 87 | CXCL11 | NA | 54059 | -0.334 | -0.1804 | Yes |
| 88 | PPBP | NA | 54235 | -0.372 | -0.1626 | Yes |
| 89 | PF4 | NA | 54448 | -0.463 | -0.1405 | Yes |
| 90 | CXCL8 | NA | 54458 | -0.470 | -0.1142 | Yes |
| 91 | FOXC1 | NA | 54472 | -0.477 | -0.0875 | Yes |
| 92 | CCR10 | NA | 54478 | -0.480 | -0.0606 | Yes |
| 93 | ACKR1 | NA | 54545 | -0.547 | -0.0311 | Yes |
| 94 | DUSP1 | NA | 54556 | -0.567 | 0.0006 | Yes |