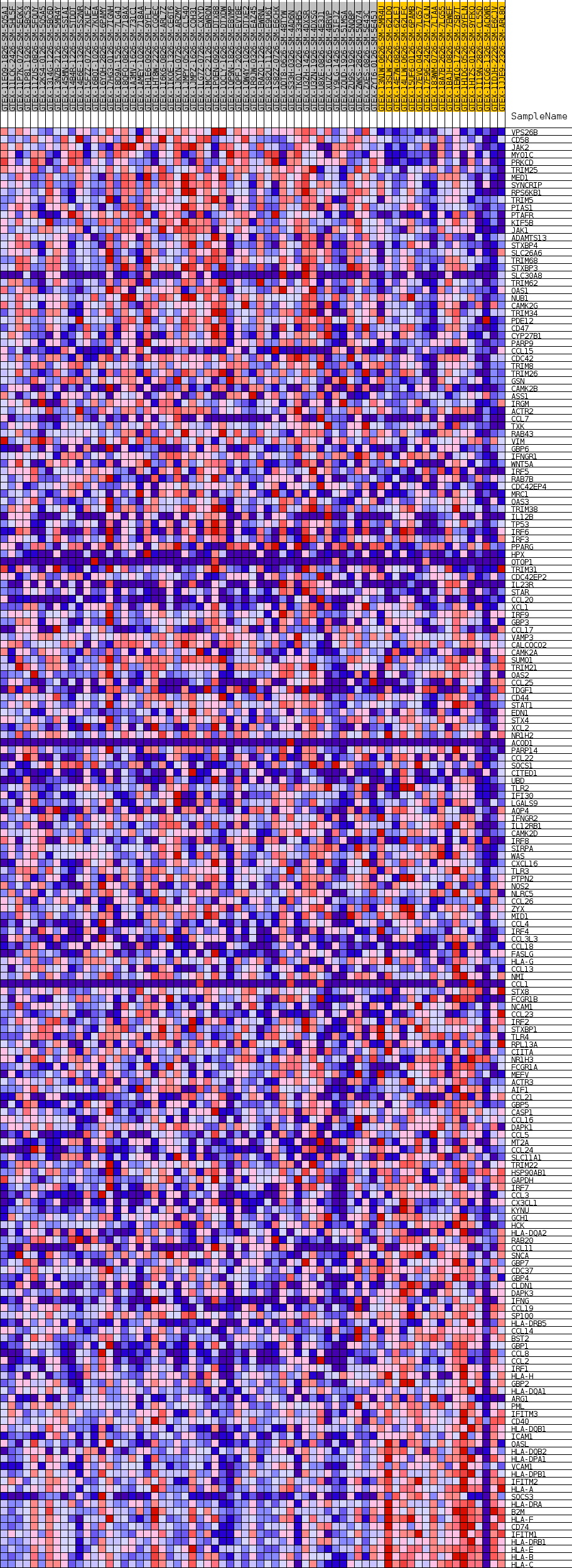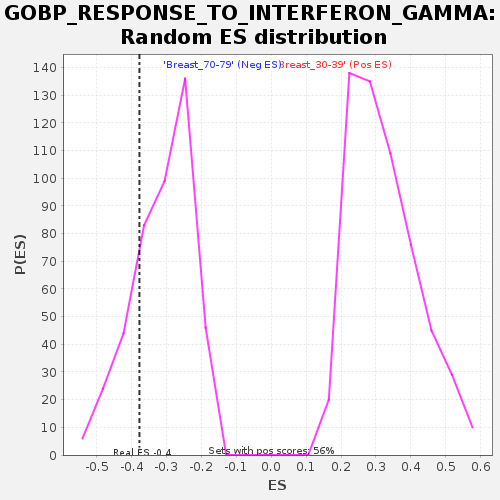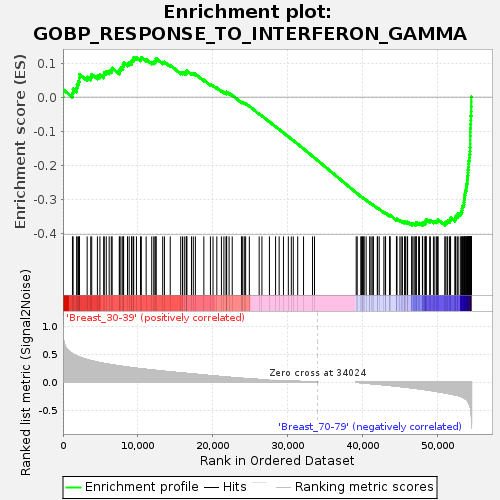
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Breast.Breast.cls #Breast_30-39_versus_Breast_70-79.Breast.cls #Breast_30-39_versus_Breast_70-79_repos |
| Phenotype | Breast.cls#Breast_30-39_versus_Breast_70-79_repos |
| Upregulated in class | Breast_70-79 |
| GeneSet | GOBP_RESPONSE_TO_INTERFERON_GAMMA |
| Enrichment Score (ES) | -0.37744018 |
| Normalized Enrichment Score (NES) | -1.2120185 |
| Nominal p-value | 0.2283105 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.998 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | VPS26B | NA | 11 | 0.876 | 0.0224 | No |
| 2 | CD58 | NA | 1264 | 0.507 | 0.0125 | No |
| 3 | JAK2 | NA | 1350 | 0.499 | 0.0239 | No |
| 4 | MYO1C | NA | 1832 | 0.466 | 0.0271 | No |
| 5 | PRKCD | NA | 1888 | 0.463 | 0.0381 | No |
| 6 | TRIM25 | NA | 2067 | 0.452 | 0.0465 | No |
| 7 | MED1 | NA | 2181 | 0.447 | 0.0560 | No |
| 8 | SYNCRIP | NA | 2206 | 0.446 | 0.0670 | No |
| 9 | RPS6KB1 | NA | 3228 | 0.401 | 0.0586 | No |
| 10 | TRIM5 | NA | 3705 | 0.384 | 0.0598 | No |
| 11 | PIAS1 | NA | 3839 | 0.379 | 0.0672 | No |
| 12 | PTAFR | NA | 4595 | 0.356 | 0.0625 | No |
| 13 | KIF5B | NA | 4934 | 0.347 | 0.0653 | No |
| 14 | JAK1 | NA | 5438 | 0.334 | 0.0646 | No |
| 15 | ADAMTS13 | NA | 5511 | 0.332 | 0.0719 | No |
| 16 | STXBP4 | NA | 5781 | 0.326 | 0.0754 | No |
| 17 | SLC26A6 | NA | 6169 | 0.317 | 0.0765 | No |
| 18 | TRIM68 | NA | 6452 | 0.310 | 0.0793 | No |
| 19 | STXBP3 | NA | 6567 | 0.308 | 0.0852 | No |
| 20 | SLC30A8 | NA | 7520 | 0.289 | 0.0751 | No |
| 21 | TRIM62 | NA | 7582 | 0.287 | 0.0815 | No |
| 22 | OAS1 | NA | 7743 | 0.284 | 0.0859 | No |
| 23 | NUB1 | NA | 8001 | 0.279 | 0.0884 | No |
| 24 | CAMK2G | NA | 8031 | 0.278 | 0.0950 | No |
| 25 | TRIM34 | NA | 8097 | 0.277 | 0.1010 | No |
| 26 | PDE12 | NA | 8619 | 0.268 | 0.0983 | No |
| 27 | CD47 | NA | 8841 | 0.264 | 0.1011 | No |
| 28 | CYP27B1 | NA | 9161 | 0.258 | 0.1019 | No |
| 29 | PARP9 | NA | 9199 | 0.258 | 0.1079 | No |
| 30 | CCL15 | NA | 9403 | 0.254 | 0.1108 | No |
| 31 | CDC42 | NA | 9447 | 0.254 | 0.1165 | No |
| 32 | TRIM8 | NA | 9813 | 0.247 | 0.1162 | No |
| 33 | TRIM26 | NA | 10352 | 0.239 | 0.1125 | No |
| 34 | GSN | NA | 10466 | 0.237 | 0.1166 | No |
| 35 | CAMK2B | NA | 11109 | 0.227 | 0.1106 | No |
| 36 | ASS1 | NA | 11856 | 0.216 | 0.1025 | No |
| 37 | IRGM | NA | 12128 | 0.213 | 0.1030 | No |
| 38 | ACTR2 | NA | 12288 | 0.211 | 0.1056 | No |
| 39 | CCL7 | NA | 12422 | 0.209 | 0.1085 | No |
| 40 | TXK | NA | 12428 | 0.209 | 0.1138 | No |
| 41 | RAB43 | NA | 13321 | 0.196 | 0.1025 | No |
| 42 | VIM | NA | 13551 | 0.193 | 0.1033 | No |
| 43 | GBP6 | NA | 14329 | 0.182 | 0.0937 | No |
| 44 | IFNGR1 | NA | 15729 | 0.165 | 0.0723 | No |
| 45 | WNT5A | NA | 15967 | 0.162 | 0.0721 | No |
| 46 | IRF5 | NA | 16205 | 0.159 | 0.0719 | No |
| 47 | RAB7B | NA | 16457 | 0.156 | 0.0713 | No |
| 48 | CDC42EP4 | NA | 16524 | 0.155 | 0.0741 | No |
| 49 | MRC1 | NA | 16542 | 0.155 | 0.0778 | No |
| 50 | OAS3 | NA | 17167 | 0.147 | 0.0701 | No |
| 51 | TRIM38 | NA | 17409 | 0.144 | 0.0694 | No |
| 52 | IL12B | NA | 17687 | 0.141 | 0.0679 | No |
| 53 | TP53 | NA | 18821 | 0.126 | 0.0504 | No |
| 54 | IRF6 | NA | 19697 | 0.115 | 0.0373 | No |
| 55 | IRF3 | NA | 20044 | 0.111 | 0.0338 | No |
| 56 | PPARG | NA | 20553 | 0.105 | 0.0271 | No |
| 57 | HPX | NA | 21168 | 0.098 | 0.0184 | No |
| 58 | OTOP1 | NA | 21500 | 0.094 | 0.0147 | No |
| 59 | TRIM31 | NA | 21806 | 0.090 | 0.0114 | No |
| 60 | CDC42EP2 | NA | 21813 | 0.090 | 0.0137 | No |
| 61 | IL23R | NA | 21868 | 0.090 | 0.0150 | No |
| 62 | STAR | NA | 22191 | 0.086 | 0.0113 | No |
| 63 | CCL20 | NA | 22612 | 0.081 | 0.0057 | No |
| 64 | XCL1 | NA | 23843 | 0.068 | -0.0152 | No |
| 65 | IRF9 | NA | 23920 | 0.068 | -0.0148 | No |
| 66 | GBP3 | NA | 24033 | 0.066 | -0.0151 | No |
| 67 | CCL17 | NA | 24278 | 0.064 | -0.0180 | No |
| 68 | VAMP3 | NA | 24404 | 0.063 | -0.0186 | No |
| 69 | CALCOCO2 | NA | 24897 | 0.058 | -0.0262 | No |
| 70 | CAMK2A | NA | 26210 | 0.046 | -0.0491 | No |
| 71 | SUMO1 | NA | 26578 | 0.043 | -0.0547 | No |
| 72 | TRIM21 | NA | 27579 | 0.035 | -0.0722 | No |
| 73 | OAS2 | NA | 28402 | 0.030 | -0.0865 | No |
| 74 | CCL25 | NA | 28881 | 0.027 | -0.0946 | No |
| 75 | TDGF1 | NA | 29457 | 0.023 | -0.1046 | No |
| 76 | CD44 | NA | 30107 | 0.020 | -0.1160 | No |
| 77 | STAT1 | NA | 30505 | 0.018 | -0.1229 | No |
| 78 | EDN1 | NA | 30768 | 0.016 | -0.1273 | No |
| 79 | STX4 | NA | 31359 | 0.014 | -0.1377 | No |
| 80 | XCL2 | NA | 32139 | 0.010 | -0.1518 | No |
| 81 | NR1H2 | NA | 33341 | 0.005 | -0.1737 | No |
| 82 | ACOD1 | NA | 33605 | 0.004 | -0.1785 | No |
| 83 | PARP14 | NA | 39174 | -0.001 | -0.2808 | No |
| 84 | CCL22 | NA | 39310 | -0.003 | -0.2832 | No |
| 85 | SOCS1 | NA | 39789 | -0.009 | -0.2918 | No |
| 86 | CITED1 | NA | 39898 | -0.010 | -0.2935 | No |
| 87 | UBD | NA | 39990 | -0.011 | -0.2949 | No |
| 88 | TLR2 | NA | 40032 | -0.012 | -0.2953 | No |
| 89 | IFI30 | NA | 40172 | -0.014 | -0.2975 | No |
| 90 | LGALS9 | NA | 40236 | -0.015 | -0.2983 | No |
| 91 | AQP4 | NA | 40504 | -0.018 | -0.3027 | No |
| 92 | IFNGR2 | NA | 40994 | -0.024 | -0.3111 | No |
| 93 | IL12RB1 | NA | 41014 | -0.024 | -0.3108 | No |
| 94 | CAMK2D | NA | 41267 | -0.027 | -0.3148 | No |
| 95 | IRF8 | NA | 41415 | -0.028 | -0.3167 | No |
| 96 | SIRPA | NA | 41470 | -0.029 | -0.3170 | No |
| 97 | WAS | NA | 41993 | -0.035 | -0.3257 | No |
| 98 | CXCL16 | NA | 42039 | -0.036 | -0.3256 | No |
| 99 | TLR3 | NA | 42249 | -0.039 | -0.3284 | No |
| 100 | PTPN2 | NA | 42872 | -0.047 | -0.3386 | No |
| 101 | NOS2 | NA | 43066 | -0.049 | -0.3409 | No |
| 102 | NLRC5 | NA | 43101 | -0.050 | -0.3402 | No |
| 103 | CCL26 | NA | 43644 | -0.057 | -0.3487 | No |
| 104 | ZYX | NA | 43662 | -0.057 | -0.3476 | No |
| 105 | MID1 | NA | 43702 | -0.058 | -0.3468 | No |
| 106 | CCL4 | NA | 44550 | -0.070 | -0.3605 | No |
| 107 | IRF4 | NA | 44587 | -0.070 | -0.3594 | No |
| 108 | CCL3L3 | NA | 44612 | -0.071 | -0.3580 | No |
| 109 | CCL18 | NA | 45013 | -0.077 | -0.3634 | No |
| 110 | FASLG | NA | 45180 | -0.079 | -0.3644 | No |
| 111 | HLA-G | NA | 45353 | -0.081 | -0.3654 | No |
| 112 | CCL13 | NA | 45642 | -0.086 | -0.3685 | No |
| 113 | NMI | NA | 45682 | -0.086 | -0.3670 | No |
| 114 | CCL1 | NA | 45784 | -0.088 | -0.3666 | No |
| 115 | STX8 | NA | 46000 | -0.091 | -0.3682 | No |
| 116 | FCGR1B | NA | 46046 | -0.092 | -0.3666 | No |
| 117 | NCAM1 | NA | 46597 | -0.101 | -0.3741 | No |
| 118 | CCL23 | NA | 46617 | -0.101 | -0.3719 | No |
| 119 | IRF2 | NA | 46774 | -0.104 | -0.3720 | No |
| 120 | STXBP1 | NA | 47007 | -0.108 | -0.3735 | No |
| 121 | TLR4 | NA | 47149 | -0.110 | -0.3733 | No |
| 122 | RPL13A | NA | 47187 | -0.111 | -0.3711 | No |
| 123 | CIITA | NA | 47192 | -0.111 | -0.3683 | No |
| 124 | NR1H3 | NA | 47515 | -0.117 | -0.3712 | No |
| 125 | FCGR1A | NA | 47631 | -0.119 | -0.3702 | No |
| 126 | MEFV | NA | 48024 | -0.126 | -0.3742 | Yes |
| 127 | ACTR3 | NA | 48033 | -0.126 | -0.3711 | Yes |
| 128 | AIF1 | NA | 48055 | -0.127 | -0.3682 | Yes |
| 129 | CCL21 | NA | 48347 | -0.132 | -0.3701 | Yes |
| 130 | GBP5 | NA | 48445 | -0.134 | -0.3684 | Yes |
| 131 | CASP1 | NA | 48465 | -0.134 | -0.3653 | Yes |
| 132 | CCL16 | NA | 48475 | -0.135 | -0.3620 | Yes |
| 133 | DAPK1 | NA | 48532 | -0.135 | -0.3595 | Yes |
| 134 | CCL5 | NA | 48995 | -0.145 | -0.3643 | Yes |
| 135 | MT2A | NA | 49083 | -0.146 | -0.3621 | Yes |
| 136 | CCL24 | NA | 49494 | -0.155 | -0.3656 | Yes |
| 137 | SLC11A1 | NA | 49682 | -0.159 | -0.3649 | Yes |
| 138 | TRIM22 | NA | 49920 | -0.164 | -0.3650 | Yes |
| 139 | HSP90AB1 | NA | 50052 | -0.168 | -0.3631 | Yes |
| 140 | GAPDH | NA | 50120 | -0.169 | -0.3600 | Yes |
| 141 | IRF7 | NA | 51030 | -0.190 | -0.3718 | Yes |
| 142 | CCL3 | NA | 51058 | -0.191 | -0.3673 | Yes |
| 143 | CX3CL1 | NA | 51304 | -0.197 | -0.3667 | Yes |
| 144 | KYNU | NA | 51397 | -0.199 | -0.3633 | Yes |
| 145 | GCH1 | NA | 51656 | -0.207 | -0.3627 | Yes |
| 146 | HCK | NA | 51729 | -0.208 | -0.3586 | Yes |
| 147 | HLA-DQA2 | NA | 51788 | -0.210 | -0.3543 | Yes |
| 148 | RAB20 | NA | 52352 | -0.225 | -0.3588 | Yes |
| 149 | CCL11 | NA | 52457 | -0.227 | -0.3548 | Yes |
| 150 | SNCA | NA | 52490 | -0.228 | -0.3495 | Yes |
| 151 | GBP7 | NA | 52757 | -0.236 | -0.3483 | Yes |
| 152 | CDC37 | NA | 52769 | -0.237 | -0.3424 | Yes |
| 153 | GBP4 | NA | 53083 | -0.249 | -0.3417 | Yes |
| 154 | CLDN1 | NA | 53215 | -0.256 | -0.3375 | Yes |
| 155 | DAPK3 | NA | 53299 | -0.261 | -0.3323 | Yes |
| 156 | IFNG | NA | 53327 | -0.263 | -0.3260 | Yes |
| 157 | CCL19 | NA | 53370 | -0.266 | -0.3199 | Yes |
| 158 | SP100 | NA | 53535 | -0.278 | -0.3157 | Yes |
| 159 | HLA-DRB5 | NA | 53577 | -0.281 | -0.3092 | Yes |
| 160 | CCL14 | NA | 53600 | -0.283 | -0.3023 | Yes |
| 161 | BST2 | NA | 53631 | -0.285 | -0.2954 | Yes |
| 162 | GBP1 | NA | 53660 | -0.288 | -0.2885 | Yes |
| 163 | CCL8 | NA | 53728 | -0.293 | -0.2821 | Yes |
| 164 | CCL2 | NA | 53749 | -0.296 | -0.2749 | Yes |
| 165 | IRF1 | NA | 53876 | -0.309 | -0.2692 | Yes |
| 166 | HLA-H | NA | 53882 | -0.309 | -0.2613 | Yes |
| 167 | GBP2 | NA | 53939 | -0.316 | -0.2541 | Yes |
| 168 | HLA-DQA1 | NA | 54024 | -0.329 | -0.2472 | Yes |
| 169 | ARG1 | NA | 54035 | -0.331 | -0.2388 | Yes |
| 170 | PML | NA | 54066 | -0.335 | -0.2307 | Yes |
| 171 | IFITM3 | NA | 54111 | -0.345 | -0.2226 | Yes |
| 172 | CD40 | NA | 54122 | -0.346 | -0.2138 | Yes |
| 173 | HLA-DQB1 | NA | 54169 | -0.356 | -0.2055 | Yes |
| 174 | ICAM1 | NA | 54181 | -0.359 | -0.1964 | Yes |
| 175 | OASL | NA | 54211 | -0.366 | -0.1875 | Yes |
| 176 | HLA-DQB2 | NA | 54298 | -0.393 | -0.1789 | Yes |
| 177 | HLA-DPA1 | NA | 54310 | -0.397 | -0.1688 | Yes |
| 178 | VCAM1 | NA | 54378 | -0.423 | -0.1591 | Yes |
| 179 | HLA-DPB1 | NA | 54388 | -0.429 | -0.1482 | Yes |
| 180 | IFITM2 | NA | 54412 | -0.438 | -0.1373 | Yes |
| 181 | HLA-A | NA | 54414 | -0.439 | -0.1259 | Yes |
| 182 | SOCS3 | NA | 54415 | -0.440 | -0.1146 | Yes |
| 183 | HLA-DRA | NA | 54422 | -0.444 | -0.1032 | Yes |
| 184 | B2M | NA | 54426 | -0.445 | -0.0917 | Yes |
| 185 | HLA-F | NA | 54455 | -0.467 | -0.0802 | Yes |
| 186 | CD74 | NA | 54486 | -0.487 | -0.0681 | Yes |
| 187 | IFITM1 | NA | 54497 | -0.495 | -0.0555 | Yes |
| 188 | HLA-DRB1 | NA | 54537 | -0.541 | -0.0422 | Yes |
| 189 | HLA-E | NA | 54547 | -0.553 | -0.0281 | Yes |
| 190 | HLA-B | NA | 54552 | -0.557 | -0.0138 | Yes |
| 191 | HLA-C | NA | 54554 | -0.559 | 0.0007 | Yes |