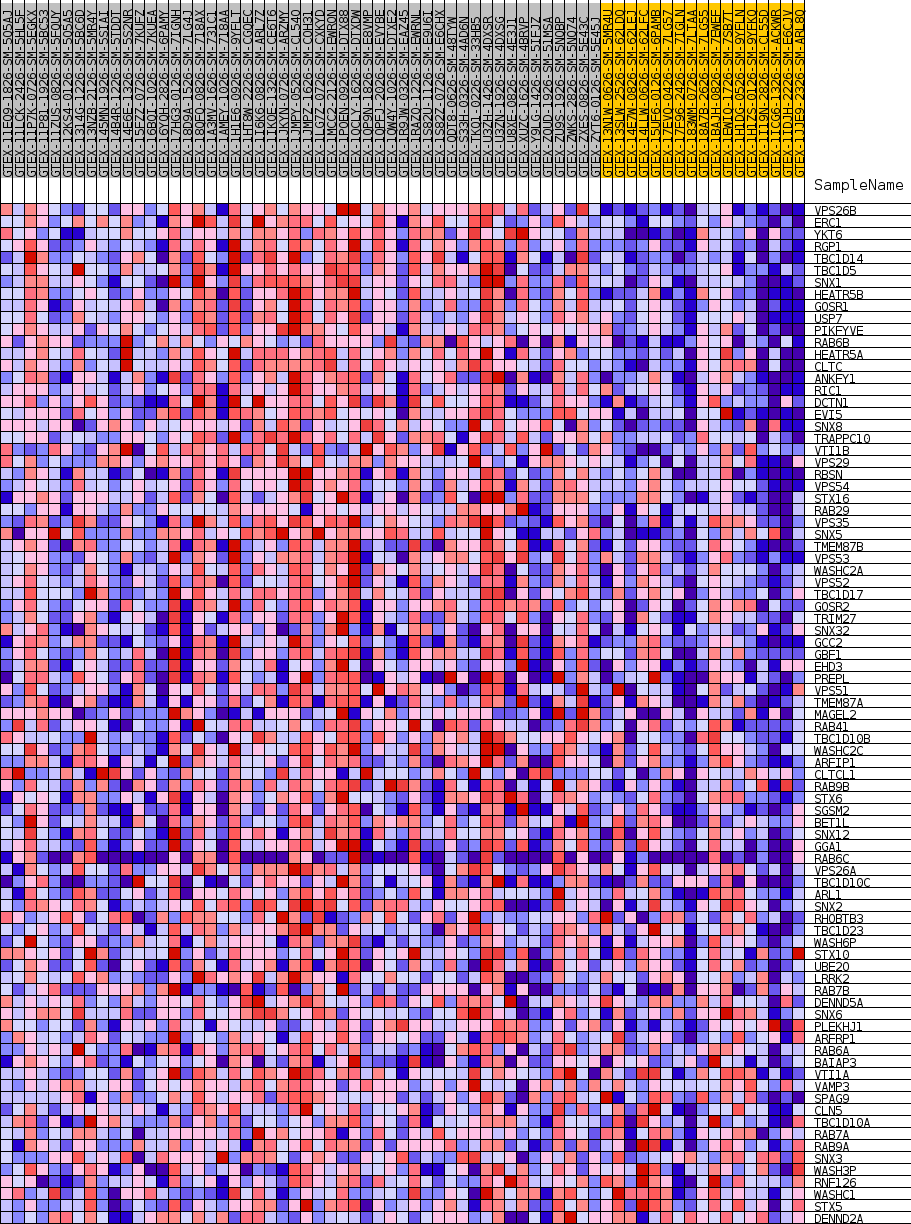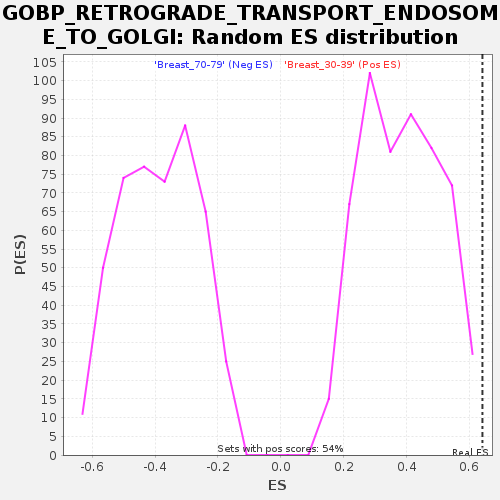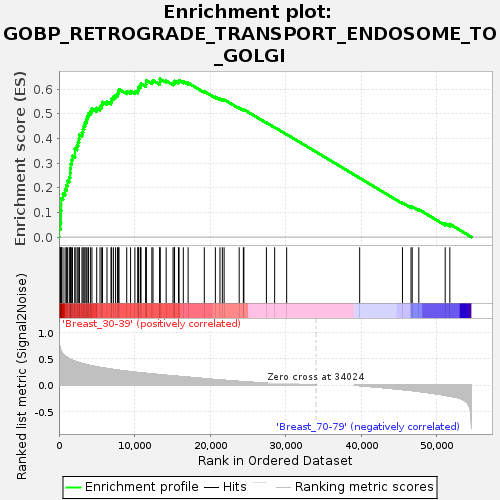
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Breast.Breast.cls #Breast_30-39_versus_Breast_70-79.Breast.cls #Breast_30-39_versus_Breast_70-79_repos |
| Phenotype | Breast.cls#Breast_30-39_versus_Breast_70-79_repos |
| Upregulated in class | Breast_30-39 |
| GeneSet | GOBP_RETROGRADE_TRANSPORT_ENDOSOME_TO_GOLGI |
| Enrichment Score (ES) | 0.6412475 |
| Normalized Enrichment Score (NES) | 1.6692116 |
| Nominal p-value | 0.0037243948 |
| FDR q-value | 0.24442409 |
| FWER p-Value | 0.754 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | VPS26B | NA | 11 | 0.876 | 0.0341 | Yes |
| 2 | ERC1 | NA | 202 | 0.672 | 0.0570 | Yes |
| 3 | YKT6 | NA | 257 | 0.652 | 0.0815 | Yes |
| 4 | RGP1 | NA | 259 | 0.652 | 0.1070 | Yes |
| 5 | TBC1D14 | NA | 297 | 0.639 | 0.1314 | Yes |
| 6 | TBC1D5 | NA | 299 | 0.638 | 0.1564 | Yes |
| 7 | SNX1 | NA | 530 | 0.588 | 0.1752 | Yes |
| 8 | HEATR5B | NA | 806 | 0.550 | 0.1917 | Yes |
| 9 | GOSR1 | NA | 981 | 0.532 | 0.2093 | Yes |
| 10 | USP7 | NA | 1111 | 0.520 | 0.2273 | Yes |
| 11 | PIKFYVE | NA | 1353 | 0.499 | 0.2425 | Yes |
| 12 | RAB6B | NA | 1476 | 0.490 | 0.2594 | Yes |
| 13 | HEATR5A | NA | 1502 | 0.488 | 0.2781 | Yes |
| 14 | CLTC | NA | 1540 | 0.486 | 0.2964 | Yes |
| 15 | ANKFY1 | NA | 1678 | 0.477 | 0.3126 | Yes |
| 16 | RIC1 | NA | 1782 | 0.469 | 0.3291 | Yes |
| 17 | DCTN1 | NA | 2112 | 0.450 | 0.3407 | Yes |
| 18 | EVI5 | NA | 2115 | 0.450 | 0.3583 | Yes |
| 19 | SNX8 | NA | 2387 | 0.436 | 0.3704 | Yes |
| 20 | TRAPPC10 | NA | 2516 | 0.430 | 0.3849 | Yes |
| 21 | VTI1B | NA | 2653 | 0.424 | 0.3990 | Yes |
| 22 | VPS29 | NA | 2699 | 0.422 | 0.4147 | Yes |
| 23 | RBSN | NA | 3041 | 0.408 | 0.4244 | Yes |
| 24 | VPS54 | NA | 3178 | 0.403 | 0.4377 | Yes |
| 25 | STX16 | NA | 3308 | 0.398 | 0.4510 | Yes |
| 26 | RAB29 | NA | 3442 | 0.393 | 0.4639 | Yes |
| 27 | VPS35 | NA | 3595 | 0.388 | 0.4763 | Yes |
| 28 | SNX5 | NA | 3720 | 0.383 | 0.4891 | Yes |
| 29 | TMEM87B | NA | 3902 | 0.377 | 0.5005 | Yes |
| 30 | VPS53 | NA | 4159 | 0.369 | 0.5103 | Yes |
| 31 | WASHC2A | NA | 4360 | 0.363 | 0.5208 | Yes |
| 32 | VPS52 | NA | 4991 | 0.346 | 0.5228 | Yes |
| 33 | TBC1D17 | NA | 5431 | 0.334 | 0.5278 | Yes |
| 34 | GOSR2 | NA | 5659 | 0.329 | 0.5365 | Yes |
| 35 | TRIM27 | NA | 5760 | 0.326 | 0.5475 | Yes |
| 36 | SNX32 | NA | 6351 | 0.313 | 0.5489 | Yes |
| 37 | GCC2 | NA | 6886 | 0.301 | 0.5509 | Yes |
| 38 | GBF1 | NA | 6966 | 0.300 | 0.5612 | Yes |
| 39 | EHD3 | NA | 7201 | 0.295 | 0.5685 | Yes |
| 40 | PREPL | NA | 7490 | 0.289 | 0.5745 | Yes |
| 41 | VPS51 | NA | 7737 | 0.284 | 0.5812 | Yes |
| 42 | TMEM87A | NA | 7858 | 0.282 | 0.5900 | Yes |
| 43 | MAGEL2 | NA | 7945 | 0.280 | 0.5994 | Yes |
| 44 | RAB41 | NA | 8966 | 0.262 | 0.5909 | Yes |
| 45 | TBC1D10B | NA | 9475 | 0.253 | 0.5915 | Yes |
| 46 | WASHC2C | NA | 10047 | 0.244 | 0.5906 | Yes |
| 47 | ARFIP1 | NA | 10422 | 0.238 | 0.5931 | Yes |
| 48 | CLTCL1 | NA | 10491 | 0.237 | 0.6011 | Yes |
| 49 | RAB9B | NA | 10560 | 0.236 | 0.6091 | Yes |
| 50 | STX6 | NA | 10790 | 0.232 | 0.6139 | Yes |
| 51 | SGSM2 | NA | 10856 | 0.231 | 0.6218 | Yes |
| 52 | BET1L | NA | 11486 | 0.222 | 0.6190 | Yes |
| 53 | SNX12 | NA | 11531 | 0.221 | 0.6268 | Yes |
| 54 | GGA1 | NA | 11540 | 0.221 | 0.6353 | Yes |
| 55 | RAB6C | NA | 12293 | 0.211 | 0.6298 | Yes |
| 56 | VPS26A | NA | 12442 | 0.209 | 0.6352 | Yes |
| 57 | TBC1D10C | NA | 13328 | 0.196 | 0.6267 | Yes |
| 58 | ARL1 | NA | 13370 | 0.195 | 0.6336 | Yes |
| 59 | SNX2 | NA | 13371 | 0.195 | 0.6412 | Yes |
| 60 | RHOBTB3 | NA | 14184 | 0.184 | 0.6336 | No |
| 61 | TBC1D23 | NA | 15095 | 0.173 | 0.6236 | No |
| 62 | WASH6P | NA | 15266 | 0.171 | 0.6272 | No |
| 63 | STX10 | NA | 15295 | 0.170 | 0.6333 | No |
| 64 | UBE2O | NA | 15816 | 0.164 | 0.6302 | No |
| 65 | LRRK2 | NA | 15895 | 0.163 | 0.6351 | No |
| 66 | RAB7B | NA | 16457 | 0.156 | 0.6310 | No |
| 67 | DENND5A | NA | 17092 | 0.148 | 0.6251 | No |
| 68 | SNX6 | NA | 19235 | 0.121 | 0.5906 | No |
| 69 | PLEKHJ1 | NA | 20699 | 0.103 | 0.5678 | No |
| 70 | ARFRP1 | NA | 21309 | 0.096 | 0.5603 | No |
| 71 | RAB6A | NA | 21622 | 0.092 | 0.5582 | No |
| 72 | BAIAP3 | NA | 21858 | 0.090 | 0.5574 | No |
| 73 | VTI1A | NA | 23852 | 0.068 | 0.5236 | No |
| 74 | VAMP3 | NA | 24404 | 0.063 | 0.5159 | No |
| 75 | SPAG9 | NA | 24476 | 0.062 | 0.5170 | No |
| 76 | CLN5 | NA | 27457 | 0.036 | 0.4638 | No |
| 77 | TBC1D10A | NA | 28547 | 0.029 | 0.4450 | No |
| 78 | RAB7A | NA | 30138 | 0.020 | 0.4165 | No |
| 79 | RAB9A | NA | 39802 | -0.009 | 0.2396 | No |
| 80 | SNX3 | NA | 45467 | -0.083 | 0.1390 | No |
| 81 | WASH3P | NA | 46600 | -0.101 | 0.1221 | No |
| 82 | RNF126 | NA | 46754 | -0.103 | 0.1234 | No |
| 83 | WASHC1 | NA | 47632 | -0.119 | 0.1119 | No |
| 84 | STX5 | NA | 51123 | -0.192 | 0.0554 | No |
| 85 | DENND2A | NA | 51741 | -0.208 | 0.0523 | No |