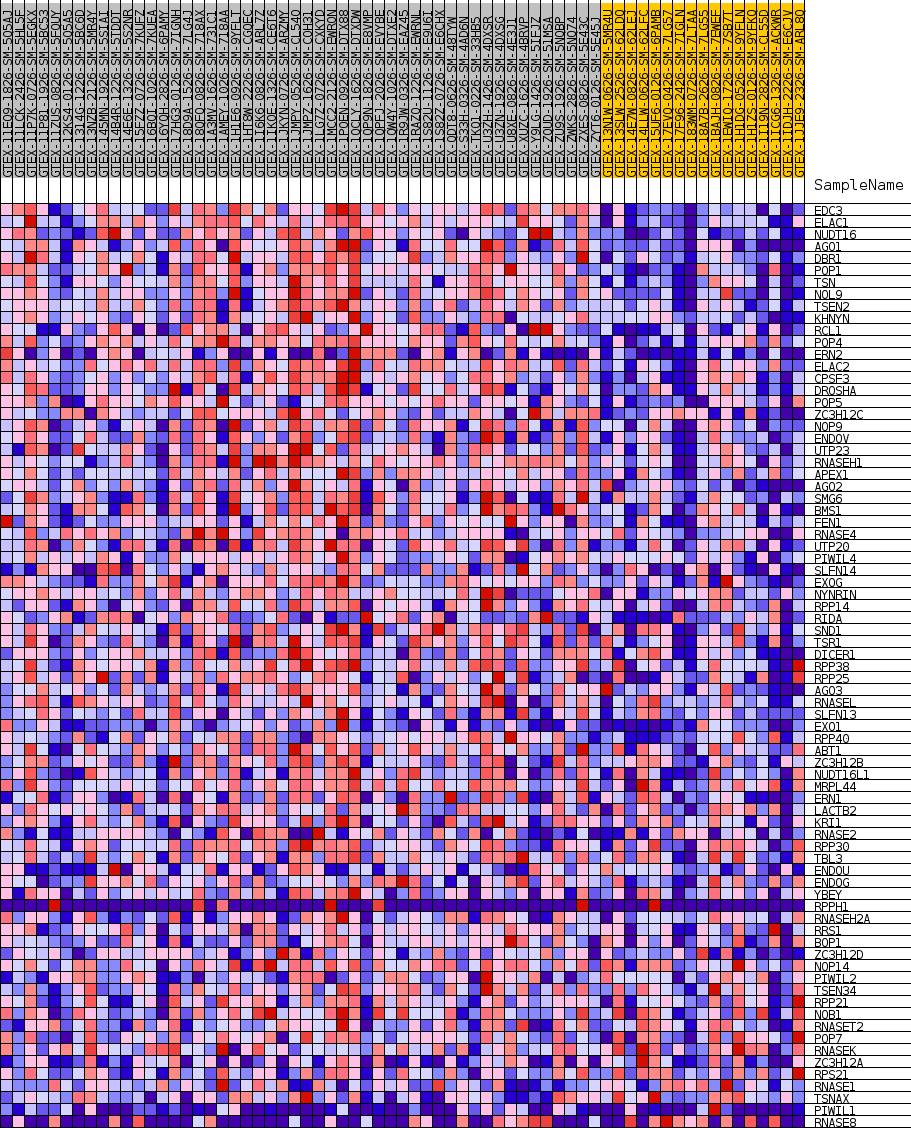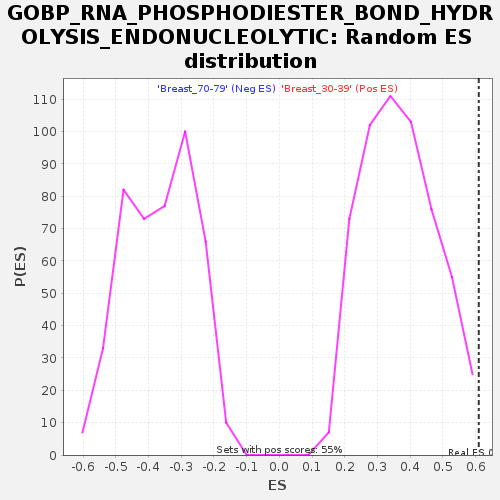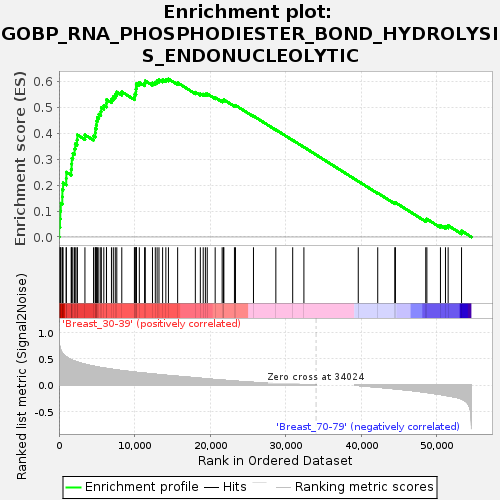
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Breast.Breast.cls #Breast_30-39_versus_Breast_70-79.Breast.cls #Breast_30-39_versus_Breast_70-79_repos |
| Phenotype | Breast.cls#Breast_30-39_versus_Breast_70-79_repos |
| Upregulated in class | Breast_30-39 |
| GeneSet | GOBP_RNA_PHOSPHODIESTER_BOND_HYDROLYSIS_ENDONUCLEOLYTIC |
| Enrichment Score (ES) | 0.60955036 |
| Normalized Enrichment Score (NES) | 1.6581827 |
| Nominal p-value | 0.0036231885 |
| FDR q-value | 0.23923558 |
| FWER p-Value | 0.785 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | EDC3 | NA | 22 | 0.842 | 0.0387 | Yes |
| 2 | ELAC1 | NA | 127 | 0.707 | 0.0697 | Yes |
| 3 | NUDT16 | NA | 171 | 0.687 | 0.1008 | Yes |
| 4 | AGO1 | NA | 235 | 0.658 | 0.1302 | Yes |
| 5 | DBR1 | NA | 435 | 0.606 | 0.1547 | Yes |
| 6 | POP1 | NA | 446 | 0.605 | 0.1826 | Yes |
| 7 | TSN | NA | 537 | 0.587 | 0.2083 | Yes |
| 8 | NOL9 | NA | 953 | 0.535 | 0.2255 | Yes |
| 9 | TSEN2 | NA | 968 | 0.533 | 0.2500 | Yes |
| 10 | KHNYN | NA | 1611 | 0.481 | 0.2606 | Yes |
| 11 | RCL1 | NA | 1664 | 0.477 | 0.2819 | Yes |
| 12 | POP4 | NA | 1695 | 0.476 | 0.3034 | Yes |
| 13 | ERN2 | NA | 1835 | 0.466 | 0.3225 | Yes |
| 14 | ELAC2 | NA | 2059 | 0.453 | 0.3395 | Yes |
| 15 | CPSF3 | NA | 2147 | 0.449 | 0.3587 | Yes |
| 16 | DROSHA | NA | 2389 | 0.436 | 0.3746 | Yes |
| 17 | POP5 | NA | 2430 | 0.434 | 0.3940 | Yes |
| 18 | ZC3H12C | NA | 3433 | 0.393 | 0.3939 | Yes |
| 19 | NOP9 | NA | 4571 | 0.357 | 0.3896 | Yes |
| 20 | ENDOV | NA | 4800 | 0.351 | 0.4018 | Yes |
| 21 | UTP23 | NA | 4813 | 0.350 | 0.4178 | Yes |
| 22 | RNASEH1 | NA | 4922 | 0.348 | 0.4320 | Yes |
| 23 | APEX1 | NA | 4986 | 0.346 | 0.4469 | Yes |
| 24 | AGO2 | NA | 5086 | 0.344 | 0.4610 | Yes |
| 25 | SMG6 | NA | 5272 | 0.338 | 0.4734 | Yes |
| 26 | BMS1 | NA | 5535 | 0.331 | 0.4839 | Yes |
| 27 | FEN1 | NA | 5594 | 0.330 | 0.4982 | Yes |
| 28 | RNASE4 | NA | 5933 | 0.322 | 0.5070 | Yes |
| 29 | UTP20 | NA | 6283 | 0.314 | 0.5152 | Yes |
| 30 | PIWIL4 | NA | 6290 | 0.314 | 0.5297 | Yes |
| 31 | SLFN14 | NA | 6944 | 0.300 | 0.5316 | Yes |
| 32 | EXOG | NA | 7189 | 0.295 | 0.5409 | Yes |
| 33 | NYNRIN | NA | 7448 | 0.290 | 0.5496 | Yes |
| 34 | RPP14 | NA | 7652 | 0.286 | 0.5592 | Yes |
| 35 | RIDA | NA | 8311 | 0.273 | 0.5598 | Yes |
| 36 | SND1 | NA | 9982 | 0.245 | 0.5406 | Yes |
| 37 | TSR1 | NA | 10039 | 0.244 | 0.5509 | Yes |
| 38 | DICER1 | NA | 10185 | 0.242 | 0.5594 | Yes |
| 39 | RPP38 | NA | 10190 | 0.241 | 0.5706 | Yes |
| 40 | RPP25 | NA | 10240 | 0.241 | 0.5809 | Yes |
| 41 | AGO3 | NA | 10258 | 0.240 | 0.5917 | Yes |
| 42 | RNASEL | NA | 10634 | 0.234 | 0.5957 | Yes |
| 43 | SLFN13 | NA | 11328 | 0.224 | 0.5934 | Yes |
| 44 | EXO1 | NA | 11422 | 0.223 | 0.6021 | Yes |
| 45 | RPP40 | NA | 12375 | 0.210 | 0.5944 | Yes |
| 46 | ABT1 | NA | 12747 | 0.205 | 0.5971 | Yes |
| 47 | ZC3H12B | NA | 12987 | 0.201 | 0.6020 | Yes |
| 48 | NUDT16L1 | NA | 13235 | 0.198 | 0.6067 | Yes |
| 49 | MRPL44 | NA | 13715 | 0.191 | 0.6067 | Yes |
| 50 | ERN1 | NA | 14146 | 0.184 | 0.6074 | Yes |
| 51 | LACTB2 | NA | 14488 | 0.180 | 0.6096 | Yes |
| 52 | KRI1 | NA | 15706 | 0.165 | 0.5949 | No |
| 53 | RNASE2 | NA | 18048 | 0.136 | 0.5583 | No |
| 54 | RPP30 | NA | 18710 | 0.127 | 0.5521 | No |
| 55 | TBL3 | NA | 19091 | 0.123 | 0.5508 | No |
| 56 | ENDOU | NA | 19378 | 0.119 | 0.5511 | No |
| 57 | ENDOG | NA | 19623 | 0.116 | 0.5520 | No |
| 58 | YBEY | NA | 20674 | 0.103 | 0.5375 | No |
| 59 | RPPH1 | NA | 21598 | 0.093 | 0.5249 | No |
| 60 | RNASEH2A | NA | 21766 | 0.091 | 0.5261 | No |
| 61 | RRS1 | NA | 21825 | 0.090 | 0.5292 | No |
| 62 | BOP1 | NA | 23225 | 0.075 | 0.5070 | No |
| 63 | ZC3H12D | NA | 23361 | 0.073 | 0.5079 | No |
| 64 | NOP14 | NA | 25744 | 0.050 | 0.4666 | No |
| 65 | PIWIL2 | NA | 28706 | 0.028 | 0.4135 | No |
| 66 | TSEN34 | NA | 30936 | 0.016 | 0.3734 | No |
| 67 | RPP21 | NA | 32420 | 0.009 | 0.3466 | No |
| 68 | NOB1 | NA | 39616 | -0.007 | 0.2149 | No |
| 69 | RNASET2 | NA | 42193 | -0.038 | 0.1694 | No |
| 70 | POP7 | NA | 44450 | -0.068 | 0.1312 | No |
| 71 | RNASEK | NA | 44537 | -0.070 | 0.1329 | No |
| 72 | ZC3H12A | NA | 48550 | -0.136 | 0.0656 | No |
| 73 | RPS21 | NA | 48699 | -0.139 | 0.0693 | No |
| 74 | RNASE1 | NA | 50496 | -0.177 | 0.0446 | No |
| 75 | TSNAX | NA | 51162 | -0.193 | 0.0414 | No |
| 76 | PIWIL1 | NA | 51514 | -0.202 | 0.0443 | No |
| 77 | RNASE8 | NA | 53288 | -0.260 | 0.0239 | No |