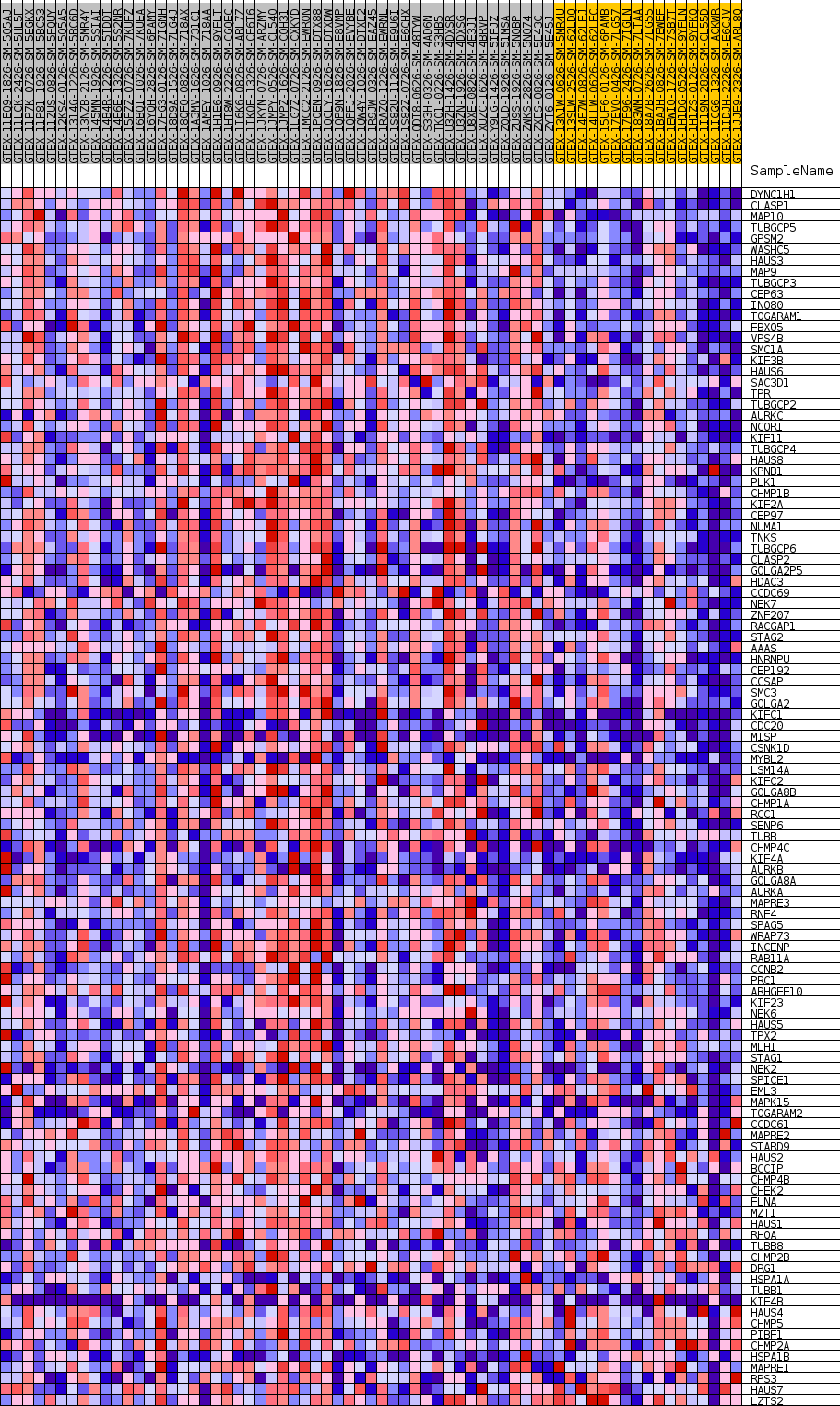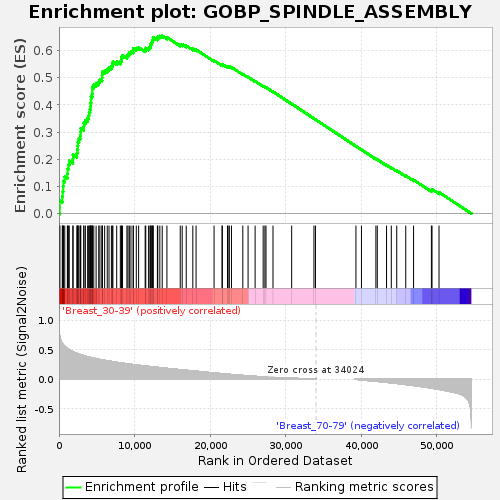
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Breast.Breast.cls #Breast_30-39_versus_Breast_70-79.Breast.cls #Breast_30-39_versus_Breast_70-79_repos |
| Phenotype | Breast.cls#Breast_30-39_versus_Breast_70-79_repos |
| Upregulated in class | Breast_30-39 |
| GeneSet | GOBP_SPINDLE_ASSEMBLY |
| Enrichment Score (ES) | 0.65414655 |
| Normalized Enrichment Score (NES) | 1.6428483 |
| Nominal p-value | 0.0018416207 |
| FDR q-value | 0.23782478 |
| FWER p-Value | 0.824 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | DYNC1H1 | NA | 96 | 0.728 | 0.0231 | Yes |
| 2 | CLASP1 | NA | 98 | 0.728 | 0.0479 | Yes |
| 3 | MAP10 | NA | 432 | 0.607 | 0.0624 | Yes |
| 4 | TUBGCP5 | NA | 495 | 0.595 | 0.0816 | Yes |
| 5 | GPSM2 | NA | 560 | 0.584 | 0.1003 | Yes |
| 6 | WASHC5 | NA | 578 | 0.581 | 0.1198 | Yes |
| 7 | HAUS3 | NA | 762 | 0.555 | 0.1354 | Yes |
| 8 | MAP9 | NA | 1108 | 0.520 | 0.1468 | Yes |
| 9 | TUBGCP3 | NA | 1144 | 0.517 | 0.1638 | Yes |
| 10 | CEP63 | NA | 1241 | 0.509 | 0.1793 | Yes |
| 11 | INO80 | NA | 1359 | 0.498 | 0.1942 | Yes |
| 12 | TOGARAM1 | NA | 1841 | 0.466 | 0.2012 | Yes |
| 13 | FBXO5 | NA | 1857 | 0.465 | 0.2168 | Yes |
| 14 | VPS4B | NA | 2362 | 0.438 | 0.2225 | Yes |
| 15 | SMC1A | NA | 2437 | 0.434 | 0.2359 | Yes |
| 16 | KIF3B | NA | 2494 | 0.431 | 0.2496 | Yes |
| 17 | HAUS6 | NA | 2513 | 0.430 | 0.2639 | Yes |
| 18 | SAC3D1 | NA | 2639 | 0.425 | 0.2761 | Yes |
| 19 | TPR | NA | 2817 | 0.417 | 0.2871 | Yes |
| 20 | TUBGCP2 | NA | 2857 | 0.415 | 0.3005 | Yes |
| 21 | AURKC | NA | 2880 | 0.413 | 0.3142 | Yes |
| 22 | NCOR1 | NA | 3282 | 0.399 | 0.3204 | Yes |
| 23 | KIF11 | NA | 3291 | 0.399 | 0.3339 | Yes |
| 24 | TUBGCP4 | NA | 3491 | 0.391 | 0.3436 | Yes |
| 25 | HAUS8 | NA | 3790 | 0.380 | 0.3511 | Yes |
| 26 | KPNB1 | NA | 3930 | 0.376 | 0.3613 | Yes |
| 27 | PLK1 | NA | 3995 | 0.374 | 0.3729 | Yes |
| 28 | CHMP1B | NA | 4091 | 0.371 | 0.3838 | Yes |
| 29 | KIF2A | NA | 4175 | 0.369 | 0.3949 | Yes |
| 30 | CEP97 | NA | 4185 | 0.369 | 0.4073 | Yes |
| 31 | NUMA1 | NA | 4237 | 0.367 | 0.4189 | Yes |
| 32 | TNKS | NA | 4245 | 0.367 | 0.4313 | Yes |
| 33 | TUBGCP6 | NA | 4381 | 0.363 | 0.4411 | Yes |
| 34 | CLASP2 | NA | 4407 | 0.362 | 0.4530 | Yes |
| 35 | GOLGA2P5 | NA | 4408 | 0.362 | 0.4654 | Yes |
| 36 | HDAC3 | NA | 4602 | 0.356 | 0.4740 | Yes |
| 37 | CCDC69 | NA | 4911 | 0.348 | 0.4802 | Yes |
| 38 | NEK7 | NA | 5261 | 0.338 | 0.4853 | Yes |
| 39 | ZNF207 | NA | 5414 | 0.334 | 0.4939 | Yes |
| 40 | RACGAP1 | NA | 5701 | 0.327 | 0.4998 | Yes |
| 41 | STAG2 | NA | 5711 | 0.327 | 0.5108 | Yes |
| 42 | AAAS | NA | 5728 | 0.327 | 0.5217 | Yes |
| 43 | HNRNPU | NA | 6042 | 0.320 | 0.5268 | Yes |
| 44 | CEP192 | NA | 6388 | 0.312 | 0.5311 | Yes |
| 45 | CCSAP | NA | 6620 | 0.307 | 0.5373 | Yes |
| 46 | SMC3 | NA | 6935 | 0.300 | 0.5418 | Yes |
| 47 | GOLGA2 | NA | 7054 | 0.298 | 0.5498 | Yes |
| 48 | KIFC1 | NA | 7135 | 0.296 | 0.5584 | Yes |
| 49 | CDC20 | NA | 7637 | 0.286 | 0.5590 | Yes |
| 50 | MISP | NA | 8119 | 0.277 | 0.5596 | Yes |
| 51 | CSNK1D | NA | 8247 | 0.275 | 0.5666 | Yes |
| 52 | MYBL2 | NA | 8265 | 0.274 | 0.5757 | Yes |
| 53 | LSM14A | NA | 8409 | 0.272 | 0.5823 | Yes |
| 54 | KIFC2 | NA | 8990 | 0.262 | 0.5806 | Yes |
| 55 | GOLGA8B | NA | 9133 | 0.259 | 0.5868 | Yes |
| 56 | CHMP1A | NA | 9313 | 0.256 | 0.5922 | Yes |
| 57 | RCC1 | NA | 9525 | 0.252 | 0.5970 | Yes |
| 58 | SENP6 | NA | 9825 | 0.247 | 0.5999 | Yes |
| 59 | TUBB | NA | 9829 | 0.247 | 0.6083 | Yes |
| 60 | CHMP4C | NA | 10232 | 0.241 | 0.6091 | Yes |
| 61 | KIF4A | NA | 10542 | 0.236 | 0.6115 | Yes |
| 62 | AURKB | NA | 11405 | 0.223 | 0.6033 | Yes |
| 63 | GOLGA8A | NA | 11487 | 0.222 | 0.6093 | Yes |
| 64 | AURKA | NA | 11860 | 0.216 | 0.6099 | Yes |
| 65 | MAPRE3 | NA | 12037 | 0.214 | 0.6140 | Yes |
| 66 | RNF4 | NA | 12140 | 0.213 | 0.6193 | Yes |
| 67 | SPAG5 | NA | 12165 | 0.213 | 0.6261 | Yes |
| 68 | WRAP73 | NA | 12281 | 0.211 | 0.6312 | Yes |
| 69 | INCENP | NA | 12361 | 0.210 | 0.6369 | Yes |
| 70 | RAB11A | NA | 12447 | 0.209 | 0.6425 | Yes |
| 71 | CCNB2 | NA | 12459 | 0.208 | 0.6494 | Yes |
| 72 | PRC1 | NA | 13024 | 0.200 | 0.6459 | Yes |
| 73 | ARHGEF10 | NA | 13079 | 0.200 | 0.6517 | Yes |
| 74 | KIF23 | NA | 13349 | 0.196 | 0.6534 | Yes |
| 75 | NEK6 | NA | 13666 | 0.191 | 0.6541 | Yes |
| 76 | HAUS5 | NA | 14286 | 0.183 | 0.6490 | No |
| 77 | TPX2 | NA | 16050 | 0.161 | 0.6221 | No |
| 78 | MLH1 | NA | 16308 | 0.158 | 0.6228 | No |
| 79 | STAG1 | NA | 16843 | 0.151 | 0.6181 | No |
| 80 | NEK2 | NA | 17715 | 0.140 | 0.6069 | No |
| 81 | SPICE1 | NA | 18146 | 0.135 | 0.6036 | No |
| 82 | EML3 | NA | 20527 | 0.105 | 0.5635 | No |
| 83 | MAPK15 | NA | 21586 | 0.093 | 0.5473 | No |
| 84 | TOGARAM2 | NA | 21619 | 0.092 | 0.5498 | No |
| 85 | CCDC61 | NA | 22303 | 0.085 | 0.5402 | No |
| 86 | MAPRE2 | NA | 22385 | 0.084 | 0.5416 | No |
| 87 | STARD9 | NA | 22550 | 0.082 | 0.5414 | No |
| 88 | HAUS2 | NA | 22831 | 0.079 | 0.5389 | No |
| 89 | BCCIP | NA | 24333 | 0.064 | 0.5136 | No |
| 90 | CHMP4B | NA | 25030 | 0.057 | 0.5027 | No |
| 91 | CHEK2 | NA | 25961 | 0.048 | 0.4873 | No |
| 92 | FLNA | NA | 27028 | 0.039 | 0.4691 | No |
| 93 | MZT1 | NA | 27255 | 0.037 | 0.4662 | No |
| 94 | HAUS1 | NA | 27401 | 0.037 | 0.4648 | No |
| 95 | RHOA | NA | 28328 | 0.030 | 0.4488 | No |
| 96 | TUBB8 | NA | 30800 | 0.016 | 0.4040 | No |
| 97 | CHMP2B | NA | 33749 | 0.003 | 0.3500 | No |
| 98 | DRG1 | NA | 33950 | 0.001 | 0.3464 | No |
| 99 | HSPA1A | NA | 39308 | -0.003 | 0.2481 | No |
| 100 | TUBB1 | NA | 40039 | -0.012 | 0.2351 | No |
| 101 | KIF4B | NA | 41950 | -0.035 | 0.2013 | No |
| 102 | HAUS4 | NA | 42136 | -0.037 | 0.1991 | No |
| 103 | CHMP5 | NA | 43360 | -0.053 | 0.1785 | No |
| 104 | PIBF1 | NA | 43989 | -0.062 | 0.1691 | No |
| 105 | CHMP2A | NA | 44709 | -0.072 | 0.1583 | No |
| 106 | HSPA1B | NA | 45902 | -0.090 | 0.1395 | No |
| 107 | MAPRE1 | NA | 46938 | -0.106 | 0.1242 | No |
| 108 | RPS3 | NA | 49297 | -0.151 | 0.0860 | No |
| 109 | HAUS7 | NA | 49380 | -0.153 | 0.0897 | No |
| 110 | LZTS2 | NA | 50306 | -0.173 | 0.0786 | No |