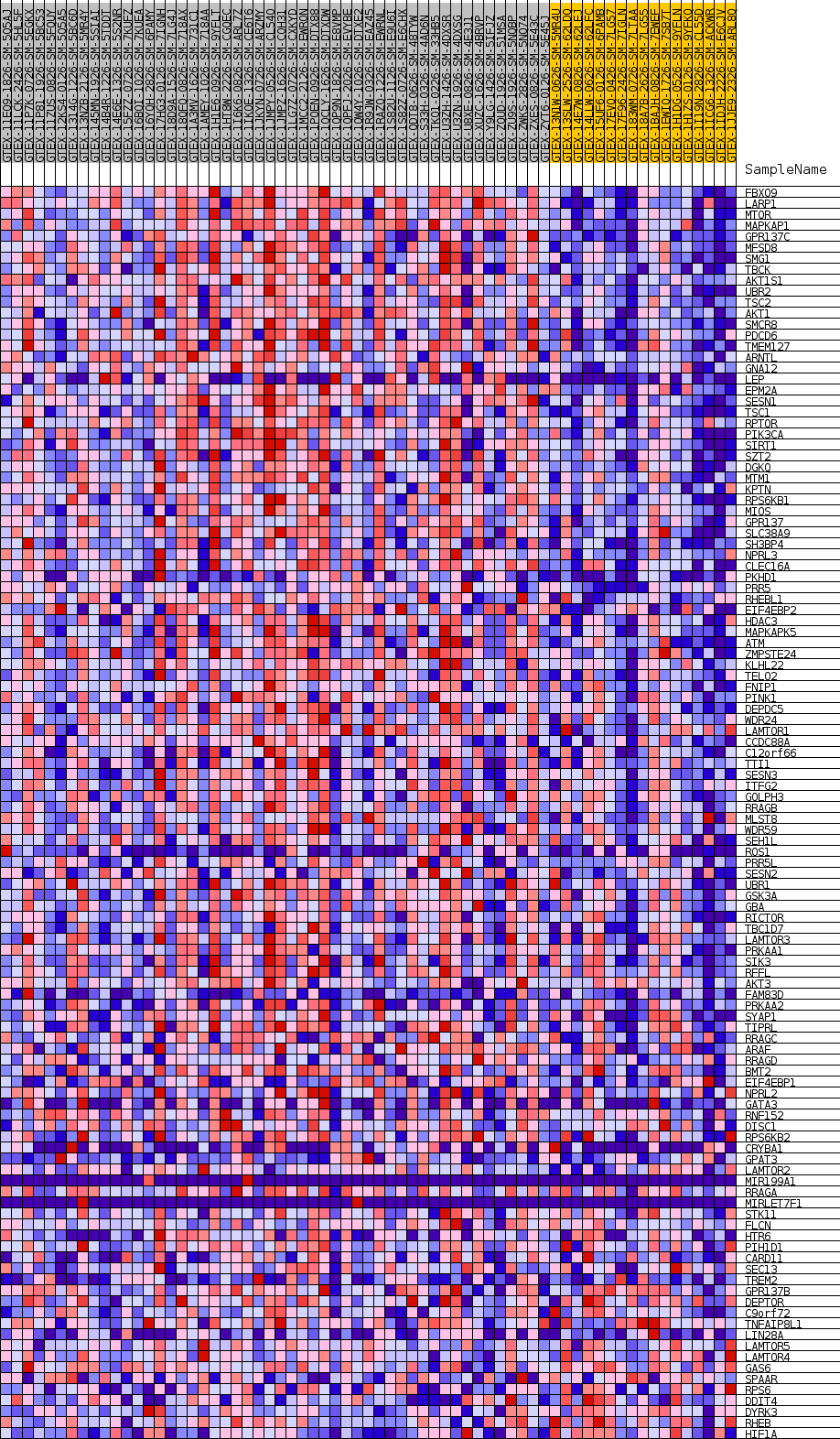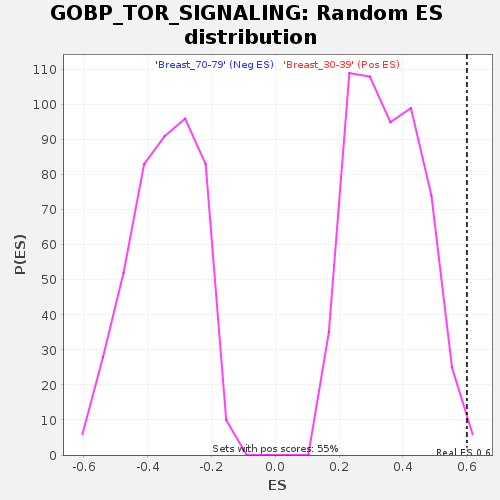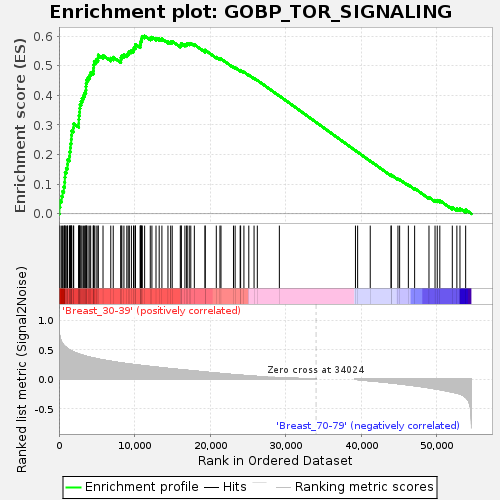
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Breast.Breast.cls #Breast_30-39_versus_Breast_70-79.Breast.cls #Breast_30-39_versus_Breast_70-79_repos |
| Phenotype | Breast.cls#Breast_30-39_versus_Breast_70-79_repos |
| Upregulated in class | Breast_30-39 |
| GeneSet | GOBP_TOR_SIGNALING |
| Enrichment Score (ES) | 0.6007384 |
| Normalized Enrichment Score (NES) | 1.7183608 |
| Nominal p-value | 0.010889292 |
| FDR q-value | 0.38733938 |
| FWER p-Value | 0.591 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | FBXO9 | NA | 53 | 0.766 | 0.0231 | Yes |
| 2 | LARP1 | NA | 129 | 0.706 | 0.0439 | Yes |
| 3 | MTOR | NA | 355 | 0.623 | 0.0593 | Yes |
| 4 | MAPKAP1 | NA | 479 | 0.597 | 0.0758 | Yes |
| 5 | GPR137C | NA | 640 | 0.571 | 0.0908 | Yes |
| 6 | MFSD8 | NA | 744 | 0.557 | 0.1064 | Yes |
| 7 | SMG1 | NA | 771 | 0.553 | 0.1233 | Yes |
| 8 | TBCK | NA | 813 | 0.549 | 0.1398 | Yes |
| 9 | AKT1S1 | NA | 960 | 0.534 | 0.1539 | Yes |
| 10 | UBR2 | NA | 1131 | 0.518 | 0.1670 | Yes |
| 11 | TSC2 | NA | 1147 | 0.516 | 0.1830 | Yes |
| 12 | AKT1 | NA | 1390 | 0.496 | 0.1941 | Yes |
| 13 | SMCR8 | NA | 1422 | 0.494 | 0.2091 | Yes |
| 14 | PDCD6 | NA | 1497 | 0.489 | 0.2231 | Yes |
| 15 | TMEM127 | NA | 1547 | 0.485 | 0.2374 | Yes |
| 16 | ARNTL | NA | 1616 | 0.481 | 0.2513 | Yes |
| 17 | GNA12 | NA | 1640 | 0.479 | 0.2659 | Yes |
| 18 | LEP | NA | 1684 | 0.477 | 0.2801 | Yes |
| 19 | EPM2A | NA | 1902 | 0.462 | 0.2906 | Yes |
| 20 | SESN1 | NA | 1945 | 0.459 | 0.3042 | Yes |
| 21 | TSC1 | NA | 2614 | 0.426 | 0.3054 | Yes |
| 22 | RPTOR | NA | 2617 | 0.426 | 0.3187 | Yes |
| 23 | PIK3CA | NA | 2652 | 0.424 | 0.3314 | Yes |
| 24 | SIRT1 | NA | 2723 | 0.421 | 0.3433 | Yes |
| 25 | SZT2 | NA | 2741 | 0.420 | 0.3562 | Yes |
| 26 | DGKQ | NA | 2788 | 0.418 | 0.3685 | Yes |
| 27 | MTM1 | NA | 2916 | 0.412 | 0.3791 | Yes |
| 28 | KPTN | NA | 3051 | 0.407 | 0.3895 | Yes |
| 29 | RPS6KB1 | NA | 3228 | 0.401 | 0.3988 | Yes |
| 30 | MIOS | NA | 3394 | 0.395 | 0.4082 | Yes |
| 31 | GPR137 | NA | 3554 | 0.389 | 0.4175 | Yes |
| 32 | SLC38A9 | NA | 3572 | 0.389 | 0.4294 | Yes |
| 33 | SH3BP4 | NA | 3593 | 0.388 | 0.4412 | Yes |
| 34 | NPRL3 | NA | 3665 | 0.385 | 0.4520 | Yes |
| 35 | CLEC16A | NA | 3849 | 0.378 | 0.4605 | Yes |
| 36 | PKHD1 | NA | 4070 | 0.372 | 0.4682 | Yes |
| 37 | PRR5 | NA | 4195 | 0.368 | 0.4774 | Yes |
| 38 | RHEBL1 | NA | 4522 | 0.359 | 0.4827 | Yes |
| 39 | EIF4EBP2 | NA | 4590 | 0.357 | 0.4927 | Yes |
| 40 | HDAC3 | NA | 4602 | 0.356 | 0.5037 | Yes |
| 41 | MAPKAPK5 | NA | 4660 | 0.355 | 0.5138 | Yes |
| 42 | ATM | NA | 4915 | 0.348 | 0.5200 | Yes |
| 43 | ZMPSTE24 | NA | 5159 | 0.341 | 0.5263 | Yes |
| 44 | KLHL22 | NA | 5191 | 0.340 | 0.5364 | Yes |
| 45 | TELO2 | NA | 5827 | 0.324 | 0.5349 | Yes |
| 46 | FNIP1 | NA | 6853 | 0.302 | 0.5256 | Yes |
| 47 | PINK1 | NA | 7180 | 0.296 | 0.5289 | Yes |
| 48 | DEPDC5 | NA | 8185 | 0.275 | 0.5191 | Yes |
| 49 | WDR24 | NA | 8205 | 0.275 | 0.5274 | Yes |
| 50 | LAMTOR1 | NA | 8320 | 0.273 | 0.5339 | Yes |
| 51 | CCDC88A | NA | 8595 | 0.268 | 0.5373 | Yes |
| 52 | C12orf66 | NA | 8979 | 0.262 | 0.5385 | Yes |
| 53 | TTI1 | NA | 9157 | 0.258 | 0.5434 | Yes |
| 54 | SESN3 | NA | 9310 | 0.256 | 0.5486 | Yes |
| 55 | ITFG2 | NA | 9584 | 0.251 | 0.5515 | Yes |
| 56 | GOLPH3 | NA | 9864 | 0.247 | 0.5541 | Yes |
| 57 | RRAGB | NA | 9928 | 0.246 | 0.5607 | Yes |
| 58 | MLST8 | NA | 10116 | 0.243 | 0.5648 | Yes |
| 59 | WDR59 | NA | 10125 | 0.242 | 0.5723 | Yes |
| 60 | SEH1L | NA | 10749 | 0.233 | 0.5682 | Yes |
| 61 | ROS1 | NA | 10772 | 0.232 | 0.5751 | Yes |
| 62 | PRR5L | NA | 10803 | 0.232 | 0.5818 | Yes |
| 63 | SESN2 | NA | 10921 | 0.230 | 0.5869 | Yes |
| 64 | UBR1 | NA | 10962 | 0.230 | 0.5933 | Yes |
| 65 | GSK3A | NA | 11015 | 0.229 | 0.5996 | Yes |
| 66 | GBA | NA | 11336 | 0.224 | 0.6007 | Yes |
| 67 | RICTOR | NA | 12082 | 0.214 | 0.5938 | No |
| 68 | TBC1D7 | NA | 12299 | 0.211 | 0.5964 | No |
| 69 | LAMTOR3 | NA | 12841 | 0.203 | 0.5929 | No |
| 70 | PRKAA1 | NA | 13261 | 0.197 | 0.5914 | No |
| 71 | SIK3 | NA | 13622 | 0.192 | 0.5908 | No |
| 72 | RFFL | NA | 14425 | 0.181 | 0.5817 | No |
| 73 | AKT3 | NA | 14784 | 0.176 | 0.5807 | No |
| 74 | FAM83D | NA | 14987 | 0.174 | 0.5825 | No |
| 75 | PRKAA2 | NA | 16056 | 0.161 | 0.5679 | No |
| 76 | SYAP1 | NA | 16173 | 0.159 | 0.5708 | No |
| 77 | TIPRL | NA | 16177 | 0.159 | 0.5757 | No |
| 78 | RRAGC | NA | 16668 | 0.153 | 0.5715 | No |
| 79 | ARAF | NA | 16899 | 0.150 | 0.5720 | No |
| 80 | RRAGD | NA | 16964 | 0.150 | 0.5756 | No |
| 81 | BMT2 | NA | 17242 | 0.146 | 0.5751 | No |
| 82 | EIF4EBP1 | NA | 17446 | 0.144 | 0.5759 | No |
| 83 | NPRL2 | NA | 17916 | 0.138 | 0.5716 | No |
| 84 | GATA3 | NA | 19337 | 0.120 | 0.5493 | No |
| 85 | RNF152 | NA | 19344 | 0.120 | 0.5529 | No |
| 86 | DISC1 | NA | 20823 | 0.102 | 0.5290 | No |
| 87 | RPS6KB2 | NA | 21306 | 0.096 | 0.5231 | No |
| 88 | CRYBA1 | NA | 21443 | 0.094 | 0.5236 | No |
| 89 | GPAT3 | NA | 23098 | 0.076 | 0.4956 | No |
| 90 | LAMTOR2 | NA | 23317 | 0.074 | 0.4939 | No |
| 91 | MIR199A1 | NA | 23983 | 0.067 | 0.4838 | No |
| 92 | RRAGA | NA | 24043 | 0.066 | 0.4848 | No |
| 93 | MIRLET7F1 | NA | 24479 | 0.062 | 0.4788 | No |
| 94 | STK11 | NA | 25125 | 0.056 | 0.4687 | No |
| 95 | FLCN | NA | 25822 | 0.049 | 0.4575 | No |
| 96 | HTR6 | NA | 26254 | 0.046 | 0.4510 | No |
| 97 | PIH1D1 | NA | 29171 | 0.025 | 0.3983 | No |
| 98 | CARD11 | NA | 39245 | -0.002 | 0.2134 | No |
| 99 | SEC13 | NA | 39533 | -0.005 | 0.2083 | No |
| 100 | TREM2 | NA | 41202 | -0.026 | 0.1785 | No |
| 101 | GPR137B | NA | 43967 | -0.062 | 0.1297 | No |
| 102 | DEPTOR | NA | 43974 | -0.062 | 0.1316 | No |
| 103 | C9orf72 | NA | 44870 | -0.074 | 0.1175 | No |
| 104 | TNFAIP8L1 | NA | 45093 | -0.078 | 0.1158 | No |
| 105 | LIN28A | NA | 46247 | -0.095 | 0.0977 | No |
| 106 | LAMTOR5 | NA | 47078 | -0.109 | 0.0858 | No |
| 107 | LAMTOR4 | NA | 48977 | -0.144 | 0.0555 | No |
| 108 | GAS6 | NA | 49780 | -0.161 | 0.0459 | No |
| 109 | SPAAR | NA | 50072 | -0.168 | 0.0458 | No |
| 110 | RPS6 | NA | 50413 | -0.175 | 0.0451 | No |
| 111 | DDIT4 | NA | 52066 | -0.217 | 0.0216 | No |
| 112 | DYRK3 | NA | 52670 | -0.234 | 0.0178 | No |
| 113 | RHEB | NA | 53080 | -0.249 | 0.0181 | No |
| 114 | HIF1A | NA | 53833 | -0.305 | 0.0139 | No |