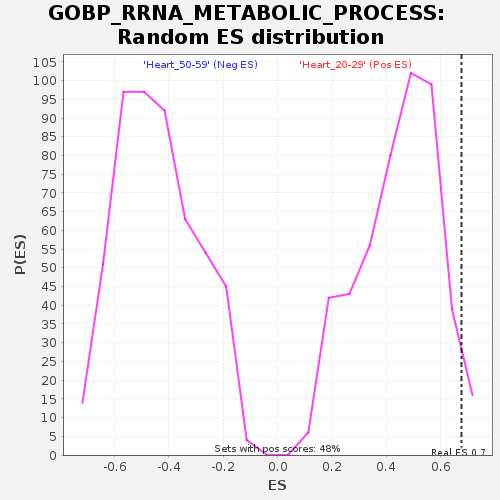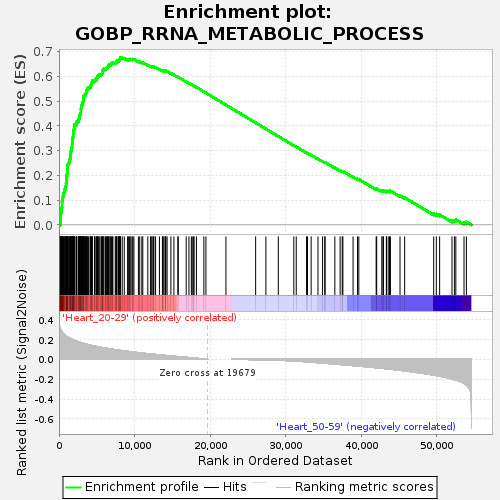
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Heart.Heart.cls #Heart_20-29_versus_Heart_50-59.Heart.cls #Heart_20-29_versus_Heart_50-59_repos |
| Phenotype | Heart.cls#Heart_20-29_versus_Heart_50-59_repos |
| Upregulated in class | Heart_20-29 |
| GeneSet | GOBP_RRNA_METABOLIC_PROCESS |
| Enrichment Score (ES) | 0.67722976 |
| Normalized Enrichment Score (NES) | 1.5198576 |
| Nominal p-value | 0.035196688 |
| FDR q-value | 0.3954227 |
| FWER p-Value | 0.766 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | RPF1 | NA | 157 | 0.309 | 0.0071 | Yes |
| 2 | MRPL1 | NA | 175 | 0.307 | 0.0167 | Yes |
| 3 | DCAF13 | NA | 198 | 0.303 | 0.0261 | Yes |
| 4 | NOP9 | NA | 212 | 0.302 | 0.0357 | Yes |
| 5 | RRP15 | NA | 241 | 0.297 | 0.0448 | Yes |
| 6 | UTP3 | NA | 274 | 0.294 | 0.0537 | Yes |
| 7 | TSR1 | NA | 333 | 0.285 | 0.0618 | Yes |
| 8 | PA2G4 | NA | 347 | 0.284 | 0.0708 | Yes |
| 9 | RPL7L1 | NA | 398 | 0.277 | 0.0788 | Yes |
| 10 | TFB2M | NA | 400 | 0.276 | 0.0877 | Yes |
| 11 | MPHOSPH10 | NA | 410 | 0.275 | 0.0964 | Yes |
| 12 | BMS1 | NA | 463 | 0.270 | 0.1042 | Yes |
| 13 | UTP15 | NA | 501 | 0.267 | 0.1122 | Yes |
| 14 | WDR3 | NA | 543 | 0.263 | 0.1199 | Yes |
| 15 | NOL11 | NA | 592 | 0.258 | 0.1274 | Yes |
| 16 | GAR1 | NA | 678 | 0.250 | 0.1339 | Yes |
| 17 | UTP11 | NA | 761 | 0.244 | 0.1403 | Yes |
| 18 | NIFK | NA | 798 | 0.242 | 0.1475 | Yes |
| 19 | RRS1 | NA | 832 | 0.241 | 0.1547 | Yes |
| 20 | SBDS | NA | 934 | 0.234 | 0.1604 | Yes |
| 21 | POP4 | NA | 943 | 0.233 | 0.1678 | Yes |
| 22 | GTPBP4 | NA | 964 | 0.232 | 0.1749 | Yes |
| 23 | WBP11 | NA | 970 | 0.232 | 0.1823 | Yes |
| 24 | EXOSC9 | NA | 992 | 0.230 | 0.1894 | Yes |
| 25 | TRMT61B | NA | 1002 | 0.230 | 0.1967 | Yes |
| 26 | RPP40 | NA | 1053 | 0.227 | 0.2031 | Yes |
| 27 | MRPS11 | NA | 1069 | 0.226 | 0.2101 | Yes |
| 28 | UTP14A | NA | 1083 | 0.226 | 0.2172 | Yes |
| 29 | EBNA1BP2 | NA | 1089 | 0.226 | 0.2244 | Yes |
| 30 | BMT2 | NA | 1109 | 0.225 | 0.2313 | Yes |
| 31 | UTP4 | NA | 1123 | 0.224 | 0.2383 | Yes |
| 32 | DDX52 | NA | 1157 | 0.222 | 0.2449 | Yes |
| 33 | BYSL | NA | 1275 | 0.217 | 0.2498 | Yes |
| 34 | METTL5 | NA | 1336 | 0.215 | 0.2556 | Yes |
| 35 | IMP4 | NA | 1434 | 0.211 | 0.2606 | Yes |
| 36 | C1D | NA | 1440 | 0.211 | 0.2674 | Yes |
| 37 | WDR12 | NA | 1490 | 0.208 | 0.2732 | Yes |
| 38 | TFB1M | NA | 1516 | 0.207 | 0.2794 | Yes |
| 39 | RPF2 | NA | 1522 | 0.207 | 0.2861 | Yes |
| 40 | KRR1 | NA | 1563 | 0.206 | 0.2920 | Yes |
| 41 | POP7 | NA | 1577 | 0.205 | 0.2984 | Yes |
| 42 | MRTO4 | NA | 1614 | 0.204 | 0.3043 | Yes |
| 43 | DKC1 | NA | 1635 | 0.203 | 0.3105 | Yes |
| 44 | RRP12 | NA | 1714 | 0.201 | 0.3156 | Yes |
| 45 | RIOK2 | NA | 1730 | 0.201 | 0.3218 | Yes |
| 46 | MAK16 | NA | 1734 | 0.201 | 0.3282 | Yes |
| 47 | RPUSD2 | NA | 1794 | 0.198 | 0.3336 | Yes |
| 48 | FRG1 | NA | 1805 | 0.198 | 0.3398 | Yes |
| 49 | CHD7 | NA | 1822 | 0.197 | 0.3459 | Yes |
| 50 | EMG1 | NA | 1839 | 0.196 | 0.3519 | Yes |
| 51 | RPS2 | NA | 1860 | 0.196 | 0.3579 | Yes |
| 52 | DIMT1 | NA | 1875 | 0.195 | 0.3640 | Yes |
| 53 | NOL10 | NA | 1882 | 0.195 | 0.3702 | Yes |
| 54 | UTP20 | NA | 1919 | 0.194 | 0.3758 | Yes |
| 55 | SHQ1 | NA | 1953 | 0.193 | 0.3814 | Yes |
| 56 | SRFBP1 | NA | 1969 | 0.192 | 0.3873 | Yes |
| 57 | RPP30 | NA | 2010 | 0.191 | 0.3928 | Yes |
| 58 | RPL7 | NA | 2011 | 0.191 | 0.3990 | Yes |
| 59 | DDX21 | NA | 2023 | 0.191 | 0.4049 | Yes |
| 60 | NOLC1 | NA | 2245 | 0.184 | 0.4068 | Yes |
| 61 | RBFA | NA | 2309 | 0.182 | 0.4116 | Yes |
| 62 | NOP14 | NA | 2315 | 0.182 | 0.4174 | Yes |
| 63 | NOL9 | NA | 2518 | 0.176 | 0.4193 | Yes |
| 64 | FCF1 | NA | 2604 | 0.174 | 0.4234 | Yes |
| 65 | ESF1 | NA | 2607 | 0.174 | 0.4290 | Yes |
| 66 | MPHOSPH6 | NA | 2686 | 0.172 | 0.4331 | Yes |
| 67 | SNU13 | NA | 2695 | 0.171 | 0.4385 | Yes |
| 68 | DDX18 | NA | 2738 | 0.170 | 0.4433 | Yes |
| 69 | PRKDC | NA | 2780 | 0.169 | 0.4480 | Yes |
| 70 | PDCD11 | NA | 2874 | 0.167 | 0.4517 | Yes |
| 71 | DIS3 | NA | 2891 | 0.167 | 0.4568 | Yes |
| 72 | XRN2 | NA | 2893 | 0.167 | 0.4622 | Yes |
| 73 | RPL5 | NA | 2894 | 0.167 | 0.4676 | Yes |
| 74 | WDR43 | NA | 2930 | 0.166 | 0.4723 | Yes |
| 75 | RRP9 | NA | 2935 | 0.166 | 0.4776 | Yes |
| 76 | NSUN4 | NA | 2985 | 0.164 | 0.4820 | Yes |
| 77 | RSL1D1 | NA | 3046 | 0.163 | 0.4862 | Yes |
| 78 | METTL15 | NA | 3089 | 0.162 | 0.4906 | Yes |
| 79 | EIF6 | NA | 3126 | 0.161 | 0.4952 | Yes |
| 80 | RPUSD4 | NA | 3169 | 0.160 | 0.4996 | Yes |
| 81 | RPUSD3 | NA | 3232 | 0.159 | 0.5036 | Yes |
| 82 | NHP2 | NA | 3247 | 0.158 | 0.5085 | Yes |
| 83 | HEATR1 | NA | 3260 | 0.158 | 0.5134 | Yes |
| 84 | RPL26 | NA | 3284 | 0.158 | 0.5180 | Yes |
| 85 | WDR74 | NA | 3293 | 0.158 | 0.5230 | Yes |
| 86 | MRM1 | NA | 3383 | 0.156 | 0.5264 | Yes |
| 87 | TBL3 | NA | 3522 | 0.153 | 0.5288 | Yes |
| 88 | UTP23 | NA | 3558 | 0.152 | 0.5331 | Yes |
| 89 | MRM2 | NA | 3601 | 0.152 | 0.5372 | Yes |
| 90 | FDXACB1 | NA | 3607 | 0.152 | 0.5420 | Yes |
| 91 | EXOSC2 | NA | 3682 | 0.150 | 0.5455 | Yes |
| 92 | BOP1 | NA | 3736 | 0.149 | 0.5494 | Yes |
| 93 | WDR46 | NA | 3802 | 0.148 | 0.5530 | Yes |
| 94 | RRP36 | NA | 3911 | 0.146 | 0.5557 | Yes |
| 95 | DDX54 | NA | 4107 | 0.142 | 0.5567 | Yes |
| 96 | MRM3 | NA | 4163 | 0.141 | 0.5603 | Yes |
| 97 | NSUN5 | NA | 4216 | 0.140 | 0.5638 | Yes |
| 98 | DIS3L | NA | 4247 | 0.140 | 0.5678 | Yes |
| 99 | NOP10 | NA | 4260 | 0.139 | 0.5721 | Yes |
| 100 | EXOSC6 | NA | 4320 | 0.138 | 0.5755 | Yes |
| 101 | EXOSC1 | NA | 4397 | 0.137 | 0.5785 | Yes |
| 102 | DROSHA | NA | 4400 | 0.137 | 0.5829 | Yes |
| 103 | NOB1 | NA | 4693 | 0.132 | 0.5818 | Yes |
| 104 | NOP56 | NA | 4748 | 0.132 | 0.5851 | Yes |
| 105 | PES1 | NA | 4765 | 0.131 | 0.5891 | Yes |
| 106 | METTL15P1 | NA | 4987 | 0.128 | 0.5891 | Yes |
| 107 | WDR18 | NA | 5011 | 0.128 | 0.5928 | Yes |
| 108 | NUDT16 | NA | 5027 | 0.127 | 0.5967 | Yes |
| 109 | NOL6 | NA | 5116 | 0.126 | 0.5991 | Yes |
| 110 | DDX10 | NA | 5120 | 0.126 | 0.6032 | Yes |
| 111 | MAPT | NA | 5256 | 0.124 | 0.6047 | Yes |
| 112 | NSA2 | NA | 5297 | 0.124 | 0.6080 | Yes |
| 113 | URB1 | NA | 5435 | 0.121 | 0.6094 | Yes |
| 114 | DDX49 | NA | 5617 | 0.119 | 0.6099 | Yes |
| 115 | RRP1B | NA | 5678 | 0.118 | 0.6126 | Yes |
| 116 | DHX37 | NA | 5741 | 0.117 | 0.6153 | Yes |
| 117 | THUMPD1 | NA | 5750 | 0.117 | 0.6189 | Yes |
| 118 | RIOK3 | NA | 5813 | 0.116 | 0.6215 | Yes |
| 119 | WDR55 | NA | 5815 | 0.116 | 0.6253 | Yes |
| 120 | FBL | NA | 5826 | 0.116 | 0.6288 | Yes |
| 121 | GTF2H5 | NA | 5953 | 0.114 | 0.6302 | Yes |
| 122 | RPS7 | NA | 6145 | 0.112 | 0.6303 | Yes |
| 123 | SENP3 | NA | 6210 | 0.111 | 0.6328 | Yes |
| 124 | RPP38 | NA | 6300 | 0.110 | 0.6347 | Yes |
| 125 | FTSJ3 | NA | 6413 | 0.109 | 0.6362 | Yes |
| 126 | NOP58 | NA | 6454 | 0.108 | 0.6389 | Yes |
| 127 | RPS24 | NA | 6520 | 0.108 | 0.6412 | Yes |
| 128 | NOP2 | NA | 6534 | 0.108 | 0.6445 | Yes |
| 129 | UTP6 | NA | 6611 | 0.106 | 0.6465 | Yes |
| 130 | RPL14 | NA | 6629 | 0.106 | 0.6496 | Yes |
| 131 | EIF4A3 | NA | 6850 | 0.103 | 0.6489 | Yes |
| 132 | EXOSC5 | NA | 6870 | 0.103 | 0.6519 | Yes |
| 133 | NAT10 | NA | 6933 | 0.102 | 0.6541 | Yes |
| 134 | SUV39H1 | NA | 7097 | 0.100 | 0.6543 | Yes |
| 135 | FAM207A | NA | 7129 | 0.100 | 0.6570 | Yes |
| 136 | RCL1 | NA | 7445 | 0.096 | 0.6543 | Yes |
| 137 | RRP8 | NA | 7533 | 0.095 | 0.6558 | Yes |
| 138 | UTP14C | NA | 7594 | 0.095 | 0.6577 | Yes |
| 139 | RRP7A | NA | 7610 | 0.094 | 0.6605 | Yes |
| 140 | EXOSC8 | NA | 7673 | 0.094 | 0.6624 | Yes |
| 141 | ABT1 | NA | 7846 | 0.092 | 0.6622 | Yes |
| 142 | RPS17 | NA | 7869 | 0.092 | 0.6648 | Yes |
| 143 | WDR36 | NA | 7989 | 0.090 | 0.6655 | Yes |
| 144 | DDX47 | NA | 7992 | 0.090 | 0.6684 | Yes |
| 145 | ERI3 | NA | 8012 | 0.090 | 0.6710 | Yes |
| 146 | RPL27 | NA | 8096 | 0.089 | 0.6724 | Yes |
| 147 | RIOK1 | NA | 8122 | 0.089 | 0.6748 | Yes |
| 148 | RPS16 | NA | 8146 | 0.089 | 0.6772 | Yes |
| 149 | ERI1 | NA | 8369 | 0.086 | 0.6759 | No |
| 150 | WDR75 | NA | 8641 | 0.084 | 0.6737 | No |
| 151 | RPL11 | NA | 9083 | 0.079 | 0.6681 | No |
| 152 | NPM3 | NA | 9178 | 0.078 | 0.6689 | No |
| 153 | MDN1 | NA | 9307 | 0.077 | 0.6691 | No |
| 154 | RPS25 | NA | 9391 | 0.076 | 0.6700 | No |
| 155 | RPL7A | NA | 9523 | 0.075 | 0.6700 | No |
| 156 | NGDN | NA | 9681 | 0.074 | 0.6695 | No |
| 157 | EXOSC4 | NA | 9778 | 0.073 | 0.6701 | No |
| 158 | NSUN3 | NA | 9919 | 0.072 | 0.6699 | No |
| 159 | UTP18 | NA | 10511 | 0.066 | 0.6611 | No |
| 160 | PIH1D2 | NA | 10684 | 0.065 | 0.6601 | No |
| 161 | ZCCHC4 | NA | 10966 | 0.063 | 0.6569 | No |
| 162 | TSR2 | NA | 11093 | 0.061 | 0.6566 | No |
| 163 | YBEY | NA | 11758 | 0.056 | 0.6462 | No |
| 164 | TEX10 | NA | 12107 | 0.053 | 0.6415 | No |
| 165 | NOC4L | NA | 12287 | 0.052 | 0.6399 | No |
| 166 | RPS21 | NA | 12320 | 0.052 | 0.6410 | No |
| 167 | EXOSC7 | NA | 12538 | 0.050 | 0.6386 | No |
| 168 | RPS6 | NA | 12543 | 0.050 | 0.6402 | No |
| 169 | RPS8 | NA | 12783 | 0.048 | 0.6373 | No |
| 170 | RPL10A | NA | 13293 | 0.044 | 0.6294 | No |
| 171 | KAT2B | NA | 13691 | 0.041 | 0.6234 | No |
| 172 | ISG20L2 | NA | 13737 | 0.041 | 0.6239 | No |
| 173 | SART1 | NA | 13750 | 0.041 | 0.6250 | No |
| 174 | NAF1 | NA | 13893 | 0.040 | 0.6237 | No |
| 175 | ZNHIT3 | NA | 14067 | 0.039 | 0.6218 | No |
| 176 | IMP3 | NA | 14086 | 0.038 | 0.6227 | No |
| 177 | RPS14 | NA | 14149 | 0.038 | 0.6228 | No |
| 178 | EXOSC3 | NA | 14312 | 0.037 | 0.6210 | No |
| 179 | PELO | NA | 14812 | 0.033 | 0.6129 | No |
| 180 | TSR3 | NA | 15212 | 0.030 | 0.6065 | No |
| 181 | LYAR | NA | 15737 | 0.027 | 0.5977 | No |
| 182 | RRP1 | NA | 15787 | 0.026 | 0.5977 | No |
| 183 | RPL35A | NA | 16838 | 0.019 | 0.5790 | No |
| 184 | REXO4 | NA | 17201 | 0.016 | 0.5729 | No |
| 185 | METTL16 | NA | 17519 | 0.014 | 0.5675 | No |
| 186 | ZNHIT6 | NA | 17635 | 0.014 | 0.5658 | No |
| 187 | MTERF4 | NA | 17833 | 0.012 | 0.5626 | No |
| 188 | RPL35 | NA | 17872 | 0.012 | 0.5623 | No |
| 189 | HELQ | NA | 18188 | 0.010 | 0.5568 | No |
| 190 | RPUSD1 | NA | 19176 | 0.003 | 0.5388 | No |
| 191 | RPS28 | NA | 19451 | 0.002 | 0.5338 | No |
| 192 | RMRP | NA | 22100 | 0.000 | 0.4851 | No |
| 193 | DDX27 | NA | 26030 | -0.004 | 0.4129 | No |
| 194 | PELP1 | NA | 27388 | -0.006 | 0.3881 | No |
| 195 | LSM6 | NA | 29029 | -0.009 | 0.3583 | No |
| 196 | PIN4 | NA | 31087 | -0.017 | 0.3210 | No |
| 197 | ERCC2 | NA | 31407 | -0.018 | 0.3157 | No |
| 198 | PWP2 | NA | 32770 | -0.025 | 0.2915 | No |
| 199 | EXOSC10 | NA | 32898 | -0.026 | 0.2900 | No |
| 200 | NSUN5P2 | NA | 33377 | -0.028 | 0.2821 | No |
| 201 | RPS27 | NA | 34282 | -0.034 | 0.2666 | No |
| 202 | NOL8 | NA | 34892 | -0.038 | 0.2566 | No |
| 203 | RPS15 | NA | 35192 | -0.040 | 0.2523 | No |
| 204 | KRI1 | NA | 35218 | -0.040 | 0.2532 | No |
| 205 | DDX51 | NA | 36516 | -0.048 | 0.2309 | No |
| 206 | NVL | NA | 37215 | -0.052 | 0.2197 | No |
| 207 | RPS9 | NA | 37493 | -0.054 | 0.2164 | No |
| 208 | ERI2 | NA | 37579 | -0.055 | 0.2166 | No |
| 209 | XRN1 | NA | 38929 | -0.063 | 0.1938 | No |
| 210 | DDX56 | NA | 39518 | -0.067 | 0.1852 | No |
| 211 | SIRT7 | NA | 39691 | -0.069 | 0.1842 | No |
| 212 | ERN2 | NA | 41985 | -0.085 | 0.1448 | No |
| 213 | ISG20 | NA | 42058 | -0.086 | 0.1462 | No |
| 214 | TRMT112 | NA | 42732 | -0.091 | 0.1368 | No |
| 215 | FBLL1 | NA | 42887 | -0.092 | 0.1369 | No |
| 216 | PIH1D1 | NA | 42946 | -0.093 | 0.1389 | No |
| 217 | LAS1L | NA | 43338 | -0.096 | 0.1348 | No |
| 218 | POP5 | NA | 43347 | -0.096 | 0.1377 | No |
| 219 | GEMIN4 | NA | 43580 | -0.097 | 0.1366 | No |
| 220 | RPS19 | NA | 43747 | -0.099 | 0.1367 | No |
| 221 | SLFN14 | NA | 43873 | -0.100 | 0.1377 | No |
| 222 | RNASEL | NA | 45144 | -0.111 | 0.1179 | No |
| 223 | HELB | NA | 45758 | -0.117 | 0.1104 | No |
| 224 | DEDD2 | NA | 49603 | -0.159 | 0.0448 | No |
| 225 | USP36 | NA | 49950 | -0.164 | 0.0437 | No |
| 226 | RPP25 | NA | 50388 | -0.170 | 0.0412 | No |
| 227 | RRNAD1 | NA | 52022 | -0.198 | 0.0176 | No |
| 228 | NSUN5P1 | NA | 52343 | -0.205 | 0.0183 | No |
| 229 | DDX17 | NA | 52532 | -0.210 | 0.0216 | No |
| 230 | RRP7BP | NA | 53616 | -0.243 | 0.0096 | No |
| 231 | SLFN13 | NA | 53930 | -0.258 | 0.0122 | No |