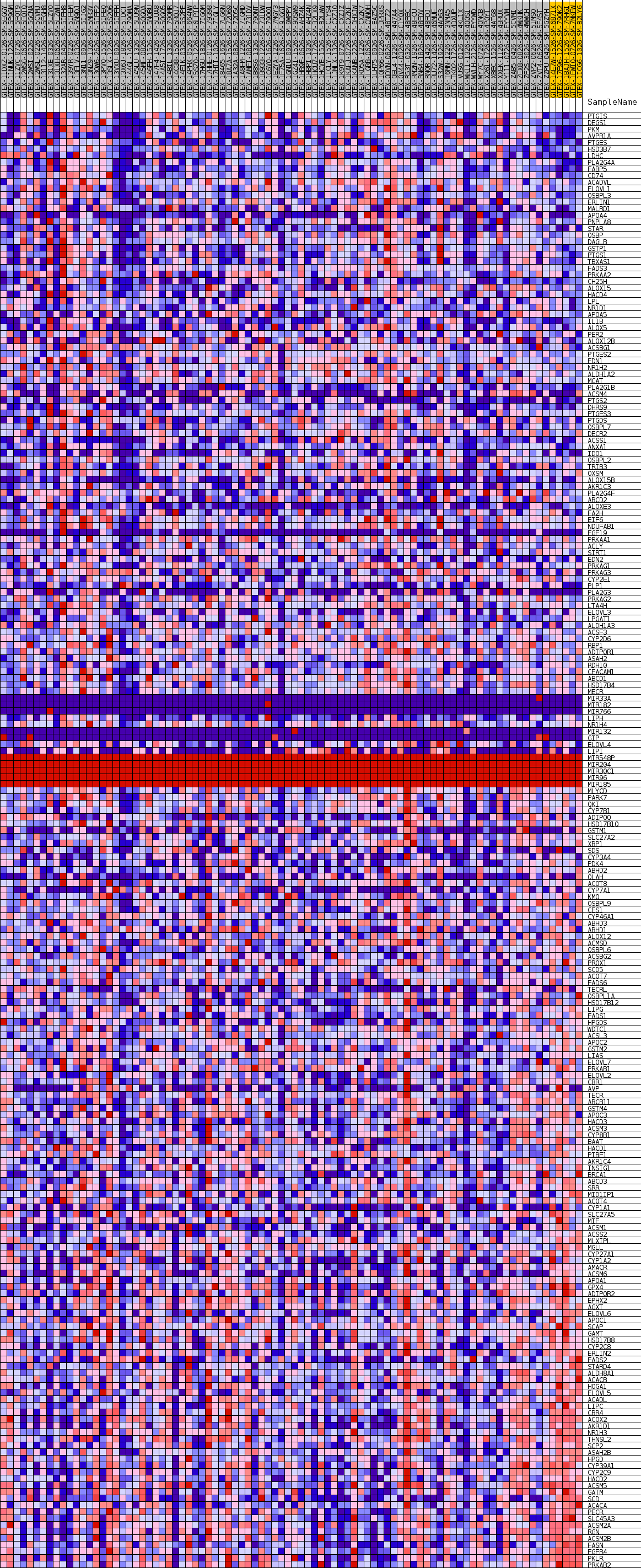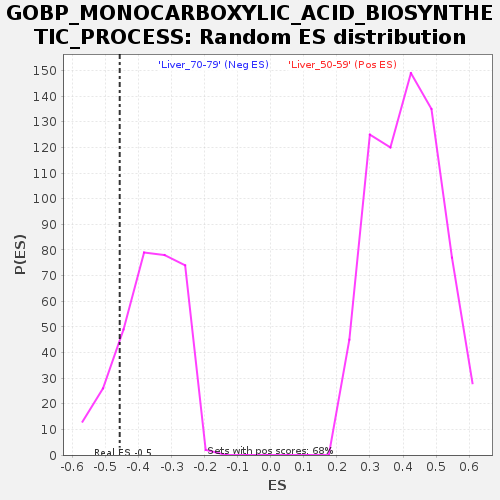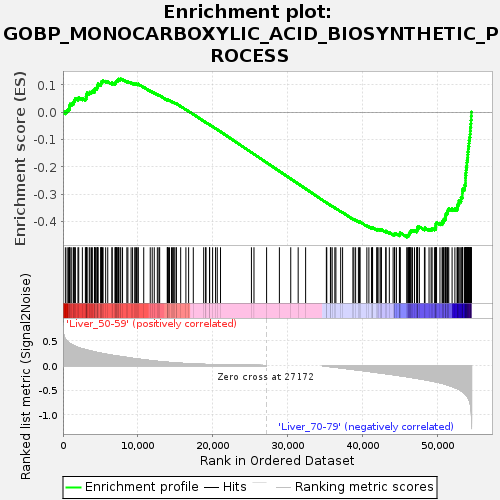
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Liver.Liver.cls #Liver_50-59_versus_Liver_70-79.Liver.cls #Liver_50-59_versus_Liver_70-79_repos |
| Phenotype | Liver.cls#Liver_50-59_versus_Liver_70-79_repos |
| Upregulated in class | Liver_70-79 |
| GeneSet | GOBP_MONOCARBOXYLIC_ACID_BIOSYNTHETIC_PROCESS |
| Enrichment Score (ES) | -0.45664945 |
| Normalized Enrichment Score (NES) | -1.2557932 |
| Nominal p-value | 0.1495327 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.929 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | PTGIS | NA | 316 | 0.534 | 0.0031 | No |
| 2 | DEGS1 | NA | 550 | 0.492 | 0.0069 | No |
| 3 | PKM | NA | 725 | 0.471 | 0.0116 | No |
| 4 | AVPR1A | NA | 884 | 0.450 | 0.0161 | No |
| 5 | PTGES | NA | 889 | 0.449 | 0.0235 | No |
| 6 | HSD3B7 | NA | 943 | 0.444 | 0.0299 | No |
| 7 | LDHC | NA | 1171 | 0.423 | 0.0327 | No |
| 8 | PLA2G4A | NA | 1395 | 0.405 | 0.0354 | No |
| 9 | FABP5 | NA | 1465 | 0.399 | 0.0407 | No |
| 10 | CD74 | NA | 1561 | 0.393 | 0.0455 | No |
| 11 | ACADVL | NA | 1655 | 0.387 | 0.0502 | No |
| 12 | ELOVL1 | NA | 2001 | 0.367 | 0.0500 | No |
| 13 | OSBPL3 | NA | 2101 | 0.362 | 0.0542 | No |
| 14 | ERLIN1 | NA | 2587 | 0.339 | 0.0509 | No |
| 15 | MALRD1 | NA | 2989 | 0.321 | 0.0488 | No |
| 16 | APOA4 | NA | 3085 | 0.317 | 0.0523 | No |
| 17 | PNPLA8 | NA | 3091 | 0.317 | 0.0575 | No |
| 18 | STAR | NA | 3145 | 0.315 | 0.0618 | No |
| 19 | OSBP | NA | 3168 | 0.314 | 0.0666 | No |
| 20 | DAGLB | NA | 3217 | 0.312 | 0.0709 | No |
| 21 | GSTP1 | NA | 3506 | 0.301 | 0.0706 | No |
| 22 | PTGS1 | NA | 3610 | 0.298 | 0.0736 | No |
| 23 | TBXAS1 | NA | 3813 | 0.290 | 0.0747 | No |
| 24 | FADS3 | NA | 3919 | 0.286 | 0.0776 | No |
| 25 | PRKAA2 | NA | 4202 | 0.277 | 0.0770 | No |
| 26 | CH25H | NA | 4206 | 0.277 | 0.0815 | No |
| 27 | ALOX15 | NA | 4250 | 0.275 | 0.0853 | No |
| 28 | HACD4 | NA | 4356 | 0.272 | 0.0879 | No |
| 29 | LPL | NA | 4536 | 0.265 | 0.0890 | No |
| 30 | NR1D1 | NA | 4568 | 0.264 | 0.0928 | No |
| 31 | APOA5 | NA | 4598 | 0.263 | 0.0966 | No |
| 32 | IL1B | NA | 4648 | 0.262 | 0.1001 | No |
| 33 | ALOX5 | NA | 4683 | 0.261 | 0.1038 | No |
| 34 | PER2 | NA | 5048 | 0.249 | 0.1012 | No |
| 35 | ALOX12B | NA | 5063 | 0.248 | 0.1051 | No |
| 36 | ACSBG1 | NA | 5116 | 0.247 | 0.1082 | No |
| 37 | PTGES2 | NA | 5137 | 0.246 | 0.1119 | No |
| 38 | EDN1 | NA | 5280 | 0.242 | 0.1134 | No |
| 39 | NR1H2 | NA | 5354 | 0.240 | 0.1160 | No |
| 40 | ALDH1A2 | NA | 5699 | 0.230 | 0.1135 | No |
| 41 | MCAT | NA | 5979 | 0.223 | 0.1121 | No |
| 42 | PLA2G1B | NA | 6549 | 0.207 | 0.1050 | No |
| 43 | ACSM4 | NA | 6558 | 0.207 | 0.1083 | No |
| 44 | PTGS2 | NA | 6942 | 0.198 | 0.1046 | No |
| 45 | DHRS9 | NA | 7008 | 0.196 | 0.1066 | No |
| 46 | PTGES3 | NA | 7039 | 0.196 | 0.1093 | No |
| 47 | PTGDS | NA | 7086 | 0.195 | 0.1117 | No |
| 48 | OSBPL7 | NA | 7149 | 0.193 | 0.1138 | No |
| 49 | DECR2 | NA | 7214 | 0.191 | 0.1158 | No |
| 50 | ACSS1 | NA | 7329 | 0.189 | 0.1168 | No |
| 51 | ANXA1 | NA | 7402 | 0.187 | 0.1186 | No |
| 52 | IDO1 | NA | 7427 | 0.187 | 0.1213 | No |
| 53 | OSBPL2 | NA | 7621 | 0.183 | 0.1208 | No |
| 54 | TRIB3 | NA | 7718 | 0.181 | 0.1220 | No |
| 55 | OXSM | NA | 7955 | 0.175 | 0.1206 | No |
| 56 | ALOX15B | NA | 8534 | 0.162 | 0.1126 | No |
| 57 | AKR1C3 | NA | 8669 | 0.159 | 0.1128 | No |
| 58 | PLA2G4F | NA | 9096 | 0.150 | 0.1075 | No |
| 59 | ABCD2 | NA | 9297 | 0.145 | 0.1062 | No |
| 60 | ALOXE3 | NA | 9583 | 0.139 | 0.1033 | No |
| 61 | FA2H | NA | 9714 | 0.137 | 0.1031 | No |
| 62 | EIF6 | NA | 9824 | 0.134 | 0.1034 | No |
| 63 | NDUFAB1 | NA | 10003 | 0.131 | 0.1023 | No |
| 64 | FGF19 | NA | 10054 | 0.130 | 0.1035 | No |
| 65 | PRKAA1 | NA | 10786 | 0.116 | 0.0920 | No |
| 66 | ACLY | NA | 11656 | 0.100 | 0.0777 | No |
| 67 | SIRT1 | NA | 11920 | 0.096 | 0.0744 | No |
| 68 | EDN2 | NA | 12194 | 0.091 | 0.0709 | No |
| 69 | PRKAG1 | NA | 12604 | 0.084 | 0.0648 | No |
| 70 | PRKAG3 | NA | 12815 | 0.081 | 0.0623 | No |
| 71 | CYP2E1 | NA | 12915 | 0.080 | 0.0618 | No |
| 72 | PLP1 | NA | 13936 | 0.066 | 0.0441 | No |
| 73 | PLA2G3 | NA | 13954 | 0.066 | 0.0449 | No |
| 74 | PRKAG2 | NA | 13961 | 0.065 | 0.0459 | No |
| 75 | LTA4H | NA | 14087 | 0.064 | 0.0446 | No |
| 76 | ELOVL3 | NA | 14233 | 0.062 | 0.0430 | No |
| 77 | LPGAT1 | NA | 14495 | 0.059 | 0.0392 | No |
| 78 | ALDH1A3 | NA | 14615 | 0.057 | 0.0379 | No |
| 79 | ACSF3 | NA | 14792 | 0.055 | 0.0356 | No |
| 80 | CYP2D6 | NA | 14859 | 0.055 | 0.0353 | No |
| 81 | RBP1 | NA | 15120 | 0.051 | 0.0314 | No |
| 82 | ADIPOR1 | NA | 15138 | 0.051 | 0.0319 | No |
| 83 | ASAH2 | NA | 15724 | 0.045 | 0.0219 | No |
| 84 | RDH10 | NA | 16421 | 0.039 | 0.0098 | No |
| 85 | CEACAM1 | NA | 16785 | 0.036 | 0.0037 | No |
| 86 | ABCD1 | NA | 17409 | 0.032 | -0.0073 | No |
| 87 | HSD17B4 | NA | 18797 | 0.023 | -0.0324 | No |
| 88 | MECR | NA | 19057 | 0.022 | -0.0368 | No |
| 89 | MIR33A | NA | 19103 | 0.022 | -0.0372 | No |
| 90 | MIR182 | NA | 19577 | 0.020 | -0.0456 | No |
| 91 | MIR766 | NA | 19595 | 0.020 | -0.0456 | No |
| 92 | LIPH | NA | 19989 | 0.018 | -0.0525 | No |
| 93 | NR1H4 | NA | 20384 | 0.017 | -0.0595 | No |
| 94 | MIR132 | NA | 20611 | 0.016 | -0.0634 | No |
| 95 | GIP | NA | 21046 | 0.014 | -0.0711 | No |
| 96 | ELOVL4 | NA | 25177 | 0.005 | -0.1470 | No |
| 97 | LIPI | NA | 25517 | 0.004 | -0.1532 | No |
| 98 | MIR548P | NA | 27204 | 0.000 | -0.1842 | No |
| 99 | MIR204 | NA | 28912 | 0.000 | -0.2156 | No |
| 100 | MIR30C1 | NA | 30430 | 0.000 | -0.2435 | No |
| 101 | MIR96 | NA | 31422 | 0.000 | -0.2617 | No |
| 102 | MIR185 | NA | 32418 | 0.000 | -0.2800 | No |
| 103 | MLYCD | NA | 35188 | -0.011 | -0.3308 | No |
| 104 | PARK7 | NA | 35243 | -0.012 | -0.3316 | No |
| 105 | QKI | NA | 35732 | -0.021 | -0.3402 | No |
| 106 | CYP7B1 | NA | 35781 | -0.021 | -0.3407 | No |
| 107 | ADIPOQ | NA | 35967 | -0.025 | -0.3437 | No |
| 108 | HSD17B10 | NA | 36321 | -0.032 | -0.3497 | No |
| 109 | GSTM1 | NA | 36462 | -0.035 | -0.3517 | No |
| 110 | SLC27A2 | NA | 37101 | -0.046 | -0.3627 | No |
| 111 | XBP1 | NA | 37358 | -0.050 | -0.3665 | No |
| 112 | SDS | NA | 38737 | -0.076 | -0.3906 | No |
| 113 | CYP3A4 | NA | 38867 | -0.078 | -0.3917 | No |
| 114 | PDK4 | NA | 39098 | -0.083 | -0.3945 | No |
| 115 | ABHD2 | NA | 39471 | -0.090 | -0.3999 | No |
| 116 | OLAH | NA | 39626 | -0.092 | -0.4012 | No |
| 117 | ACOT8 | NA | 39696 | -0.093 | -0.4009 | No |
| 118 | CYP7A1 | NA | 40599 | -0.111 | -0.4156 | No |
| 119 | KMO | NA | 40871 | -0.116 | -0.4187 | No |
| 120 | OSBPL9 | NA | 41216 | -0.124 | -0.4230 | No |
| 121 | CES1 | NA | 41357 | -0.126 | -0.4235 | No |
| 122 | CYP46A1 | NA | 41361 | -0.126 | -0.4214 | No |
| 123 | ABHD3 | NA | 41915 | -0.138 | -0.4293 | No |
| 124 | ABHD1 | NA | 41995 | -0.140 | -0.4284 | No |
| 125 | ALOX12 | NA | 42099 | -0.142 | -0.4280 | No |
| 126 | ACMSD | NA | 42368 | -0.147 | -0.4304 | No |
| 127 | OSBPL6 | NA | 42493 | -0.150 | -0.4302 | No |
| 128 | ACSBG2 | NA | 42546 | -0.151 | -0.4287 | No |
| 129 | PROX1 | NA | 43119 | -0.163 | -0.4365 | No |
| 130 | SCD5 | NA | 43167 | -0.164 | -0.4346 | No |
| 131 | ACOT7 | NA | 43602 | -0.172 | -0.4398 | No |
| 132 | FADS6 | NA | 44108 | -0.182 | -0.4460 | No |
| 133 | TECRL | NA | 44303 | -0.187 | -0.4465 | No |
| 134 | OSBPL1A | NA | 44315 | -0.187 | -0.4436 | No |
| 135 | HSD17B12 | NA | 44549 | -0.192 | -0.4447 | No |
| 136 | LIPG | NA | 44951 | -0.201 | -0.4487 | No |
| 137 | FADS1 | NA | 44958 | -0.202 | -0.4455 | No |
| 138 | HPGDS | NA | 45015 | -0.203 | -0.4431 | No |
| 139 | WDTC1 | NA | 45058 | -0.204 | -0.4405 | No |
| 140 | ACSL3 | NA | 45936 | -0.223 | -0.4529 | Yes |
| 141 | APOC2 | NA | 45999 | -0.225 | -0.4504 | Yes |
| 142 | GSTM2 | NA | 46175 | -0.229 | -0.4498 | Yes |
| 143 | LIAS | NA | 46220 | -0.230 | -0.4468 | Yes |
| 144 | ELOVL7 | NA | 46260 | -0.231 | -0.4436 | Yes |
| 145 | PRKAB1 | NA | 46298 | -0.232 | -0.4405 | Yes |
| 146 | ELOVL2 | NA | 46391 | -0.234 | -0.4383 | Yes |
| 147 | CBR1 | NA | 46481 | -0.237 | -0.4360 | Yes |
| 148 | AVP | NA | 46509 | -0.237 | -0.4325 | Yes |
| 149 | TECR | NA | 46705 | -0.242 | -0.4321 | Yes |
| 150 | ABCB11 | NA | 46968 | -0.249 | -0.4328 | Yes |
| 151 | GSTM4 | NA | 47261 | -0.256 | -0.4339 | Yes |
| 152 | APOC3 | NA | 47315 | -0.258 | -0.4306 | Yes |
| 153 | HACD3 | NA | 47336 | -0.258 | -0.4267 | Yes |
| 154 | ACSM3 | NA | 47366 | -0.259 | -0.4229 | Yes |
| 155 | CYP8B1 | NA | 47396 | -0.260 | -0.4191 | Yes |
| 156 | BAAT | NA | 47619 | -0.265 | -0.4188 | Yes |
| 157 | HACD1 | NA | 48291 | -0.283 | -0.4264 | Yes |
| 158 | PIBF1 | NA | 48367 | -0.285 | -0.4231 | Yes |
| 159 | AKR1C4 | NA | 48908 | -0.300 | -0.4280 | Yes |
| 160 | INSIG1 | NA | 49183 | -0.308 | -0.4280 | Yes |
| 161 | BRCA1 | NA | 49316 | -0.312 | -0.4252 | Yes |
| 162 | ABCD3 | NA | 49629 | -0.322 | -0.4256 | Yes |
| 163 | SRR | NA | 49803 | -0.328 | -0.4233 | Yes |
| 164 | MID1IP1 | NA | 49816 | -0.329 | -0.4181 | Yes |
| 165 | ACOT4 | NA | 49824 | -0.329 | -0.4128 | Yes |
| 166 | CYP1A1 | NA | 49836 | -0.329 | -0.4075 | Yes |
| 167 | SLC27A5 | NA | 49915 | -0.332 | -0.4034 | Yes |
| 168 | MIF | NA | 50366 | -0.348 | -0.4059 | Yes |
| 169 | ACSM1 | NA | 50671 | -0.359 | -0.4055 | Yes |
| 170 | ACSS2 | NA | 50677 | -0.359 | -0.3996 | Yes |
| 171 | MLXIPL | NA | 50857 | -0.366 | -0.3969 | Yes |
| 172 | MGLL | NA | 50878 | -0.367 | -0.3911 | Yes |
| 173 | CYP27A1 | NA | 51071 | -0.376 | -0.3884 | Yes |
| 174 | CYP1A2 | NA | 51079 | -0.376 | -0.3823 | Yes |
| 175 | AMACR | NA | 51146 | -0.380 | -0.3772 | Yes |
| 176 | ACSM6 | NA | 51172 | -0.381 | -0.3713 | Yes |
| 177 | APOA1 | NA | 51321 | -0.388 | -0.3676 | Yes |
| 178 | GPX4 | NA | 51361 | -0.390 | -0.3619 | Yes |
| 179 | ADIPOR2 | NA | 51447 | -0.394 | -0.3569 | Yes |
| 180 | EPHX2 | NA | 51547 | -0.399 | -0.3521 | Yes |
| 181 | AGXT | NA | 51987 | -0.420 | -0.3532 | Yes |
| 182 | ELOVL6 | NA | 52361 | -0.445 | -0.3527 | Yes |
| 183 | APOC1 | NA | 52657 | -0.464 | -0.3504 | Yes |
| 184 | SCAP | NA | 52701 | -0.467 | -0.3434 | Yes |
| 185 | GAMT | NA | 52751 | -0.471 | -0.3365 | Yes |
| 186 | HSD17B8 | NA | 52897 | -0.481 | -0.3312 | Yes |
| 187 | CYP2C8 | NA | 52912 | -0.482 | -0.3234 | Yes |
| 188 | ERLIN2 | NA | 53173 | -0.505 | -0.3198 | Yes |
| 189 | FADS2 | NA | 53193 | -0.507 | -0.3117 | Yes |
| 190 | STARD4 | NA | 53360 | -0.523 | -0.3061 | Yes |
| 191 | ALDH8A1 | NA | 53361 | -0.524 | -0.2974 | Yes |
| 192 | ACACB | NA | 53364 | -0.524 | -0.2888 | Yes |
| 193 | HOGA1 | NA | 53415 | -0.530 | -0.2809 | Yes |
| 194 | ELOVL5 | NA | 53655 | -0.560 | -0.2760 | Yes |
| 195 | ACADL | NA | 53668 | -0.563 | -0.2668 | Yes |
| 196 | LIPC | NA | 53781 | -0.577 | -0.2593 | Yes |
| 197 | CBR4 | NA | 53784 | -0.578 | -0.2498 | Yes |
| 198 | ACOX2 | NA | 53786 | -0.578 | -0.2402 | Yes |
| 199 | AKR1D1 | NA | 53796 | -0.580 | -0.2307 | Yes |
| 200 | NR1H3 | NA | 53836 | -0.588 | -0.2217 | Yes |
| 201 | THNSL2 | NA | 53856 | -0.591 | -0.2122 | Yes |
| 202 | SCP2 | NA | 53871 | -0.594 | -0.2026 | Yes |
| 203 | ASAH2B | NA | 53959 | -0.609 | -0.1941 | Yes |
| 204 | HPGD | NA | 53968 | -0.612 | -0.1840 | Yes |
| 205 | CYP39A1 | NA | 53997 | -0.618 | -0.1743 | Yes |
| 206 | CYP2C9 | NA | 54074 | -0.635 | -0.1652 | Yes |
| 207 | HACD2 | NA | 54091 | -0.637 | -0.1549 | Yes |
| 208 | ACSM5 | NA | 54110 | -0.643 | -0.1445 | Yes |
| 209 | GATM | NA | 54148 | -0.655 | -0.1343 | Yes |
| 210 | SCD | NA | 54171 | -0.661 | -0.1237 | Yes |
| 211 | ACACA | NA | 54240 | -0.682 | -0.1137 | Yes |
| 212 | PECR | NA | 54259 | -0.689 | -0.1025 | Yes |
| 213 | SLC45A3 | NA | 54328 | -0.718 | -0.0919 | Yes |
| 214 | ACSM2A | NA | 54389 | -0.752 | -0.0805 | Yes |
| 215 | RGN | NA | 54408 | -0.764 | -0.0681 | Yes |
| 216 | ACSM2B | NA | 54438 | -0.788 | -0.0556 | Yes |
| 217 | FASN | NA | 54494 | -0.835 | -0.0427 | Yes |
| 218 | FGFR4 | NA | 54512 | -0.848 | -0.0290 | Yes |
| 219 | PKLR | NA | 54541 | -0.898 | -0.0146 | Yes |
| 220 | PRKAB2 | NA | 54557 | -0.931 | 0.0006 | Yes |