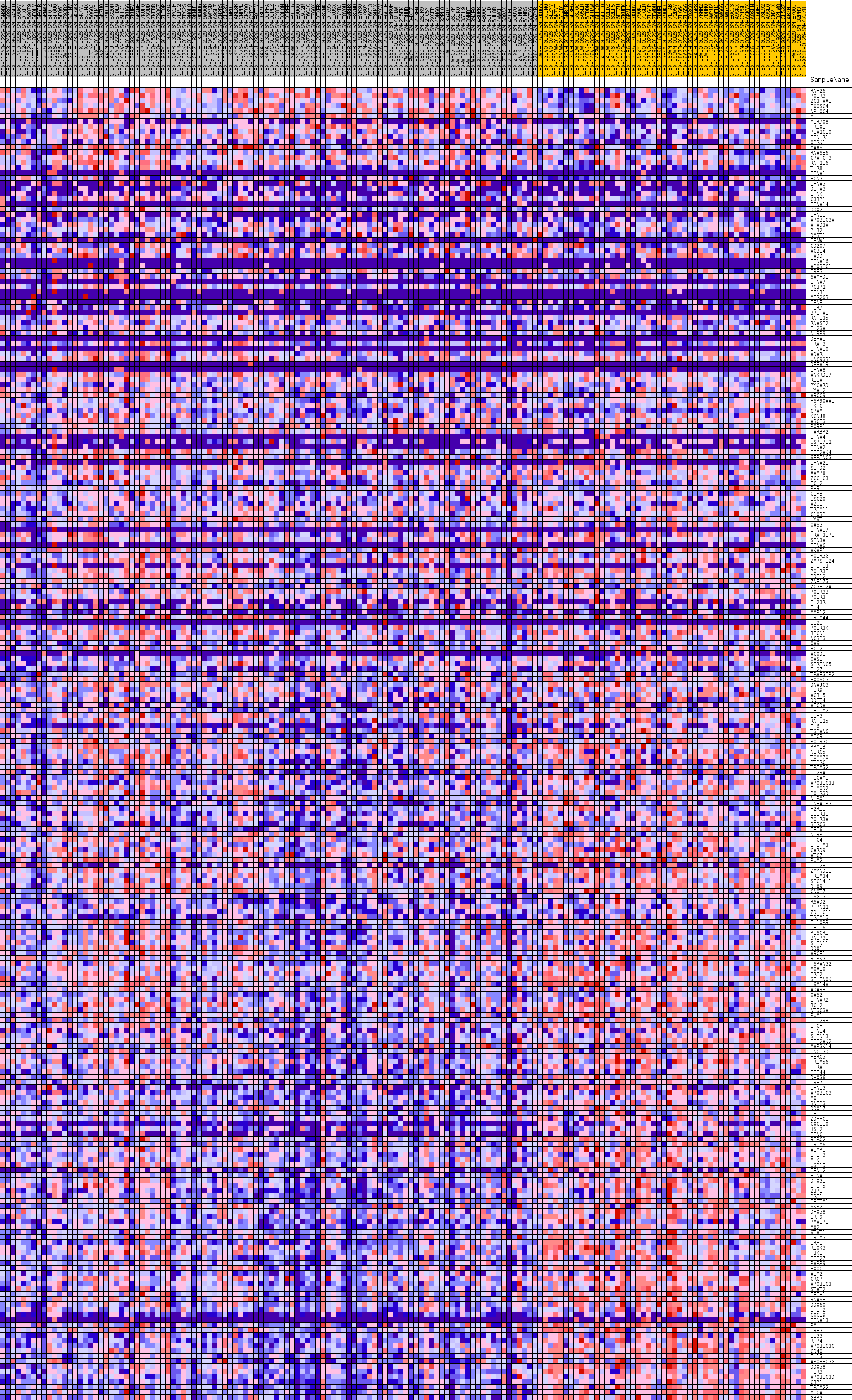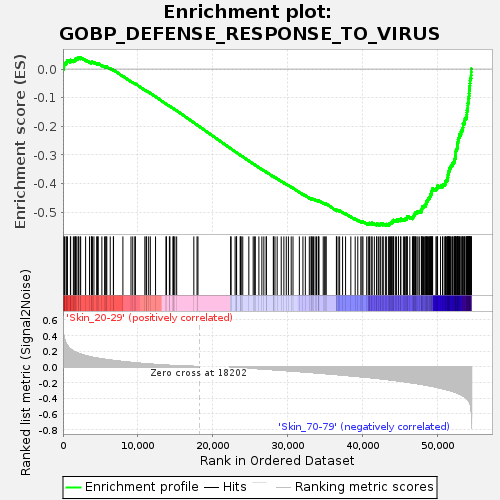
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Skin.Skin.cls #Skin_20-29_versus_Skin_70-79.Skin.cls #Skin_20-29_versus_Skin_70-79_repos |
| Phenotype | Skin.cls#Skin_20-29_versus_Skin_70-79_repos |
| Upregulated in class | Skin_70-79 |
| GeneSet | GOBP_DEFENSE_RESPONSE_TO_VIRUS |
| Enrichment Score (ES) | -0.5473387 |
| Normalized Enrichment Score (NES) | -1.7436639 |
| Nominal p-value | 0.0 |
| FDR q-value | 0.29157287 |
| FWER p-Value | 0.599 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | RNF26 | NA | 86 | 0.402 | 0.0068 | No |
| 2 | POLR3H | NA | 120 | 0.388 | 0.0142 | No |
| 3 | ZC3HAV1 | NA | 187 | 0.353 | 0.0204 | No |
| 4 | EXOSC4 | NA | 406 | 0.298 | 0.0225 | No |
| 5 | NPLOC4 | NA | 498 | 0.280 | 0.0267 | No |
| 6 | MUL1 | NA | 594 | 0.267 | 0.0305 | No |
| 7 | MIR708 | NA | 991 | 0.225 | 0.0279 | No |
| 8 | TREX1 | NA | 1022 | 0.222 | 0.0319 | No |
| 9 | PLA2G10 | NA | 1386 | 0.198 | 0.0294 | No |
| 10 | IFNLR1 | NA | 1507 | 0.192 | 0.0311 | No |
| 11 | OPRK1 | NA | 1648 | 0.186 | 0.0324 | No |
| 12 | MAVS | NA | 1660 | 0.185 | 0.0360 | No |
| 13 | RNASE6 | NA | 1773 | 0.180 | 0.0377 | No |
| 14 | GPATCH3 | NA | 2031 | 0.170 | 0.0365 | No |
| 15 | RNF216 | NA | 2038 | 0.170 | 0.0399 | No |
| 16 | TLR8 | NA | 2282 | 0.161 | 0.0388 | No |
| 17 | IFNA1 | NA | 2361 | 0.159 | 0.0407 | No |
| 18 | FCN3 | NA | 3020 | 0.139 | 0.0315 | No |
| 19 | IFNA5 | NA | 3544 | 0.127 | 0.0245 | No |
| 20 | DEFA3 | NA | 3793 | 0.122 | 0.0225 | No |
| 21 | IFNK | NA | 3805 | 0.122 | 0.0248 | No |
| 22 | G3BP1 | NA | 3933 | 0.119 | 0.0249 | No |
| 23 | IFNA14 | NA | 4134 | 0.115 | 0.0237 | No |
| 24 | DDX21 | NA | 4497 | 0.109 | 0.0193 | No |
| 25 | IFNL1 | NA | 4615 | 0.107 | 0.0193 | No |
| 26 | APOBEC3A | NA | 4708 | 0.106 | 0.0198 | No |
| 27 | ATAD3A | NA | 5194 | 0.099 | 0.0130 | No |
| 28 | PHB2 | NA | 5558 | 0.093 | 0.0082 | No |
| 29 | DMBT1 | NA | 5718 | 0.091 | 0.0072 | No |
| 30 | IFNW1 | NA | 5751 | 0.091 | 0.0085 | No |
| 31 | CD207 | NA | 5851 | 0.089 | 0.0085 | No |
| 32 | AGBL4 | NA | 6313 | 0.083 | 0.0018 | No |
| 33 | FADD | NA | 6730 | 0.078 | -0.0043 | No |
| 34 | IFNA16 | NA | 6751 | 0.078 | -0.0030 | No |
| 35 | APOBEC1 | NA | 8003 | 0.064 | -0.0247 | No |
| 36 | IRF5 | NA | 9086 | 0.053 | -0.0435 | No |
| 37 | SAMHD1 | NA | 9320 | 0.051 | -0.0467 | No |
| 38 | IFNA7 | NA | 9597 | 0.049 | -0.0508 | No |
| 39 | PCBP2 | NA | 9674 | 0.048 | -0.0512 | No |
| 40 | IFNB1 | NA | 10936 | 0.037 | -0.0736 | No |
| 41 | MIR26B | NA | 11103 | 0.036 | -0.0759 | No |
| 42 | IFNE | NA | 11175 | 0.035 | -0.0765 | No |
| 43 | TLR7 | NA | 11447 | 0.033 | -0.0808 | No |
| 44 | BPIFA1 | NA | 11668 | 0.032 | -0.0842 | No |
| 45 | RNF135 | NA | 12365 | 0.027 | -0.0964 | No |
| 46 | RNASE2 | NA | 13753 | 0.019 | -0.1216 | No |
| 47 | IL23A | NA | 13831 | 0.018 | -0.1226 | No |
| 48 | NLRP9 | NA | 14217 | 0.016 | -0.1293 | No |
| 49 | DEFA1 | NA | 14286 | 0.016 | -0.1303 | No |
| 50 | TRAF3 | NA | 14687 | 0.014 | -0.1373 | No |
| 51 | IFNA10 | NA | 14760 | 0.014 | -0.1384 | No |
| 52 | ADAR | NA | 14814 | 0.014 | -0.1391 | No |
| 53 | UNC93B1 | NA | 14949 | 0.013 | -0.1413 | No |
| 54 | DEFA1B | NA | 15188 | 0.012 | -0.1454 | No |
| 55 | IFNA8 | NA | 17479 | 0.003 | -0.1875 | No |
| 56 | ANKRD17 | NA | 17909 | 0.002 | -0.1953 | No |
| 57 | RELA | NA | 18032 | 0.001 | -0.1975 | No |
| 58 | PYCARD | NA | 22400 | -0.000 | -0.2779 | No |
| 59 | HYAL2 | NA | 22457 | -0.001 | -0.2789 | No |
| 60 | ABCC9 | NA | 22995 | -0.004 | -0.2887 | No |
| 61 | HSP90AA1 | NA | 23170 | -0.005 | -0.2918 | No |
| 62 | TKFC | NA | 23194 | -0.006 | -0.2921 | No |
| 63 | GPAM | NA | 23685 | -0.009 | -0.3009 | No |
| 64 | KCNJ8 | NA | 23698 | -0.009 | -0.3010 | No |
| 65 | ABCF3 | NA | 23814 | -0.009 | -0.3029 | No |
| 66 | PQBP1 | NA | 24018 | -0.010 | -0.3064 | No |
| 67 | TARBP2 | NA | 24830 | -0.015 | -0.3210 | No |
| 68 | IFNA4 | NA | 25409 | -0.019 | -0.3313 | No |
| 69 | USP17L2 | NA | 25566 | -0.020 | -0.3337 | No |
| 70 | IFNA2 | NA | 25696 | -0.021 | -0.3357 | No |
| 71 | EIF2AK4 | NA | 26172 | -0.024 | -0.3439 | No |
| 72 | SERINC3 | NA | 26570 | -0.026 | -0.3507 | No |
| 73 | IFNA21 | NA | 26837 | -0.028 | -0.3550 | No |
| 74 | SETD2 | NA | 27159 | -0.030 | -0.3603 | No |
| 75 | VAMP8 | NA | 27162 | -0.030 | -0.3597 | No |
| 76 | ZCCHC3 | NA | 28102 | -0.036 | -0.3762 | No |
| 77 | FGL2 | NA | 28144 | -0.037 | -0.3762 | No |
| 78 | PHB | NA | 28323 | -0.038 | -0.3787 | No |
| 79 | CLPB | NA | 28616 | -0.040 | -0.3832 | No |
| 80 | ISG20 | NA | 29159 | -0.043 | -0.3923 | No |
| 81 | AZU1 | NA | 29511 | -0.046 | -0.3978 | No |
| 82 | TRIM11 | NA | 29833 | -0.048 | -0.4027 | No |
| 83 | C1QBP | NA | 30118 | -0.050 | -0.4069 | No |
| 84 | LYST | NA | 30490 | -0.053 | -0.4127 | No |
| 85 | OAS3 | NA | 30731 | -0.054 | -0.4159 | No |
| 86 | IFNA17 | NA | 31580 | -0.060 | -0.4303 | No |
| 87 | TRAF3IP1 | NA | 32057 | -0.063 | -0.4378 | No |
| 88 | SIN3A | NA | 32379 | -0.066 | -0.4423 | No |
| 89 | IFNA6 | NA | 32909 | -0.069 | -0.4506 | No |
| 90 | AKAP1 | NA | 33138 | -0.071 | -0.4533 | No |
| 91 | POLR3G | NA | 33189 | -0.072 | -0.4527 | No |
| 92 | ZMPSTE24 | NA | 33358 | -0.073 | -0.4543 | No |
| 93 | IFIT1B | NA | 33479 | -0.074 | -0.4550 | No |
| 94 | POLR3E | NA | 33690 | -0.075 | -0.4573 | No |
| 95 | PDE12 | NA | 33875 | -0.077 | -0.4591 | No |
| 96 | ZNF175 | NA | 34110 | -0.079 | -0.4618 | No |
| 97 | ZC3H12A | NA | 34124 | -0.079 | -0.4604 | No |
| 98 | POLR3B | NA | 34203 | -0.079 | -0.4601 | No |
| 99 | POLR3F | NA | 34768 | -0.083 | -0.4688 | No |
| 100 | IL23R | NA | 34919 | -0.085 | -0.4698 | No |
| 101 | IL4 | NA | 35114 | -0.086 | -0.4716 | No |
| 102 | MMP12 | NA | 35155 | -0.087 | -0.4705 | No |
| 103 | TRIM44 | NA | 36533 | -0.098 | -0.4938 | No |
| 104 | IL21 | NA | 36539 | -0.098 | -0.4919 | No |
| 105 | POLR3K | NA | 36756 | -0.100 | -0.4938 | No |
| 106 | BECN1 | NA | 36963 | -0.101 | -0.4955 | No |
| 107 | NCBP3 | NA | 37356 | -0.105 | -0.5005 | No |
| 108 | OASL | NA | 37755 | -0.108 | -0.5056 | No |
| 109 | BCL2L1 | NA | 38458 | -0.114 | -0.5161 | No |
| 110 | ACOD1 | NA | 39040 | -0.119 | -0.5244 | No |
| 111 | OAS1 | NA | 39387 | -0.122 | -0.5282 | No |
| 112 | SERINC5 | NA | 39794 | -0.126 | -0.5330 | No |
| 113 | IL27 | NA | 39948 | -0.127 | -0.5332 | No |
| 114 | TRAF3IP2 | NA | 40076 | -0.128 | -0.5329 | No |
| 115 | EXOSC5 | NA | 40581 | -0.133 | -0.5394 | No |
| 116 | DNAJC3 | NA | 40791 | -0.135 | -0.5404 | No |
| 117 | TLR9 | NA | 40935 | -0.136 | -0.5402 | No |
| 118 | AGBL5 | NA | 41005 | -0.137 | -0.5387 | No |
| 119 | DDIT4 | NA | 41272 | -0.139 | -0.5407 | No |
| 120 | AICDA | NA | 41283 | -0.139 | -0.5379 | No |
| 121 | IFITM2 | NA | 41599 | -0.143 | -0.5408 | No |
| 122 | ILF3 | NA | 41899 | -0.146 | -0.5433 | No |
| 123 | RNF125 | NA | 41935 | -0.146 | -0.5409 | No |
| 124 | IL6 | NA | 42171 | -0.149 | -0.5421 | No |
| 125 | TSPAN6 | NA | 42361 | -0.151 | -0.5424 | No |
| 126 | MICB | NA | 42461 | -0.152 | -0.5411 | No |
| 127 | POLR3C | NA | 42720 | -0.155 | -0.5426 | No |
| 128 | PPM1B | NA | 42785 | -0.155 | -0.5406 | No |
| 129 | NLRC5 | NA | 43075 | -0.158 | -0.5426 | No |
| 130 | TOMM70 | NA | 43240 | -0.160 | -0.5423 | No |
| 131 | PTPRC | NA | 43514 | -0.163 | -0.5439 | Yes |
| 132 | TRIM52 | NA | 43567 | -0.164 | -0.5415 | Yes |
| 133 | IL2RA | NA | 43652 | -0.165 | -0.5396 | Yes |
| 134 | TICAM1 | NA | 43770 | -0.166 | -0.5383 | Yes |
| 135 | APOBEC3B | NA | 43868 | -0.167 | -0.5366 | Yes |
| 136 | ELMOD2 | NA | 43961 | -0.168 | -0.5348 | Yes |
| 137 | POLR3D | NA | 43979 | -0.169 | -0.5316 | Yes |
| 138 | NLRX1 | NA | 44148 | -0.171 | -0.5312 | Yes |
| 139 | TNFAIP3 | NA | 44177 | -0.171 | -0.5281 | Yes |
| 140 | F2RL1 | NA | 44408 | -0.174 | -0.5288 | Yes |
| 141 | LILRB1 | NA | 44551 | -0.175 | -0.5277 | Yes |
| 142 | POLR3A | NA | 44813 | -0.179 | -0.5288 | Yes |
| 143 | BIRC3 | NA | 44832 | -0.179 | -0.5254 | Yes |
| 144 | IFI6 | NA | 45122 | -0.183 | -0.5269 | Yes |
| 145 | NLRP1 | NA | 45127 | -0.183 | -0.5232 | Yes |
| 146 | TTC4 | NA | 45482 | -0.187 | -0.5259 | Yes |
| 147 | IFITM3 | NA | 45634 | -0.189 | -0.5247 | Yes |
| 148 | CARD9 | NA | 45795 | -0.191 | -0.5237 | Yes |
| 149 | ATG7 | NA | 45925 | -0.193 | -0.5220 | Yes |
| 150 | PUM2 | NA | 45972 | -0.194 | -0.5188 | Yes |
| 151 | IL12B | NA | 45995 | -0.194 | -0.5152 | Yes |
| 152 | ZMYND11 | NA | 46338 | -0.199 | -0.5174 | Yes |
| 153 | TRIM34 | NA | 46692 | -0.205 | -0.5196 | Yes |
| 154 | SEC14L1 | NA | 46774 | -0.206 | -0.5168 | Yes |
| 155 | DHX9 | NA | 46818 | -0.206 | -0.5133 | Yes |
| 156 | CNOT7 | NA | 46903 | -0.207 | -0.5106 | Yes |
| 157 | ISG15 | NA | 46935 | -0.208 | -0.5068 | Yes |
| 158 | RSAD2 | NA | 47065 | -0.210 | -0.5048 | Yes |
| 159 | PTPN22 | NA | 47102 | -0.210 | -0.5011 | Yes |
| 160 | ZDHHC11 | NA | 47279 | -0.213 | -0.5000 | Yes |
| 161 | TRIM15 | NA | 47429 | -0.215 | -0.4982 | Yes |
| 162 | IL10RB | NA | 47608 | -0.218 | -0.4970 | Yes |
| 163 | IFI16 | NA | 47877 | -0.222 | -0.4973 | Yes |
| 164 | PLSCR1 | NA | 47913 | -0.223 | -0.4933 | Yes |
| 165 | BNIP3L | NA | 47943 | -0.223 | -0.4892 | Yes |
| 166 | SLFN11 | NA | 47982 | -0.224 | -0.4853 | Yes |
| 167 | DDX1 | NA | 48008 | -0.224 | -0.4811 | Yes |
| 168 | ABCE1 | NA | 48171 | -0.227 | -0.4793 | Yes |
| 169 | RIPK3 | NA | 48324 | -0.229 | -0.4774 | Yes |
| 170 | TSPAN32 | NA | 48466 | -0.232 | -0.4751 | Yes |
| 171 | MOV10 | NA | 48501 | -0.232 | -0.4709 | Yes |
| 172 | IRF2 | NA | 48530 | -0.233 | -0.4666 | Yes |
| 173 | SELENOK | NA | 48567 | -0.234 | -0.4624 | Yes |
| 174 | LSM14A | NA | 48694 | -0.236 | -0.4598 | Yes |
| 175 | ADARB1 | NA | 48752 | -0.237 | -0.4560 | Yes |
| 176 | OAS2 | NA | 48826 | -0.238 | -0.4524 | Yes |
| 177 | IFNAR2 | NA | 48962 | -0.240 | -0.4498 | Yes |
| 178 | BCL2 | NA | 49021 | -0.242 | -0.4459 | Yes |
| 179 | NT5C3A | NA | 49078 | -0.242 | -0.4419 | Yes |
| 180 | PUM1 | NA | 49114 | -0.243 | -0.4375 | Yes |
| 181 | IL12RB1 | NA | 49241 | -0.245 | -0.4347 | Yes |
| 182 | ITCH | NA | 49266 | -0.246 | -0.4300 | Yes |
| 183 | IFNL4 | NA | 49284 | -0.246 | -0.4252 | Yes |
| 184 | SLFN13 | NA | 49364 | -0.248 | -0.4216 | Yes |
| 185 | EIF2AK2 | NA | 49387 | -0.248 | -0.4168 | Yes |
| 186 | MAP3K14 | NA | 49831 | -0.258 | -0.4196 | Yes |
| 187 | UNC13D | NA | 49937 | -0.260 | -0.4161 | Yes |
| 188 | HERC5 | NA | 50033 | -0.262 | -0.4124 | Yes |
| 189 | TRIM56 | NA | 50047 | -0.262 | -0.4072 | Yes |
| 190 | HTRA1 | NA | 50444 | -0.271 | -0.4089 | Yes |
| 191 | IFI44L | NA | 50706 | -0.277 | -0.4080 | Yes |
| 192 | DHX36 | NA | 50750 | -0.278 | -0.4030 | Yes |
| 193 | IRF7 | NA | 51005 | -0.284 | -0.4017 | Yes |
| 194 | IFNL3 | NA | 51111 | -0.286 | -0.3977 | Yes |
| 195 | APOBEC3H | NA | 51112 | -0.286 | -0.3918 | Yes |
| 196 | MX1 | NA | 51226 | -0.289 | -0.3879 | Yes |
| 197 | BNIP3 | NA | 51404 | -0.294 | -0.3850 | Yes |
| 198 | DDX17 | NA | 51415 | -0.294 | -0.3791 | Yes |
| 199 | IFIT1 | NA | 51436 | -0.295 | -0.3733 | Yes |
| 200 | ZDHHC1 | NA | 51452 | -0.296 | -0.3674 | Yes |
| 201 | CXCL10 | NA | 51482 | -0.297 | -0.3618 | Yes |
| 202 | BST2 | NA | 51541 | -0.298 | -0.3567 | Yes |
| 203 | IFNG | NA | 51638 | -0.300 | -0.3522 | Yes |
| 204 | BIRC2 | NA | 51639 | -0.300 | -0.3460 | Yes |
| 205 | TRIM6 | NA | 51786 | -0.305 | -0.3423 | Yes |
| 206 | AIMP1 | NA | 51856 | -0.307 | -0.3372 | Yes |
| 207 | IFIT3 | NA | 52023 | -0.312 | -0.3338 | Yes |
| 208 | MLKL | NA | 52070 | -0.313 | -0.3281 | Yes |
| 209 | USP15 | NA | 52232 | -0.318 | -0.3245 | Yes |
| 210 | IFNL2 | NA | 52313 | -0.321 | -0.3193 | Yes |
| 211 | FLNA | NA | 52335 | -0.322 | -0.3130 | Yes |
| 212 | DTX3L | NA | 52427 | -0.325 | -0.3079 | Yes |
| 213 | IFIT5 | NA | 52444 | -0.326 | -0.3014 | Yes |
| 214 | ZBP1 | NA | 52472 | -0.326 | -0.2951 | Yes |
| 215 | PRF1 | NA | 52475 | -0.327 | -0.2884 | Yes |
| 216 | IFITM1 | NA | 52522 | -0.328 | -0.2824 | Yes |
| 217 | SKP2 | NA | 52617 | -0.332 | -0.2772 | Yes |
| 218 | DHX58 | NA | 52703 | -0.334 | -0.2719 | Yes |
| 219 | IRF9 | NA | 52712 | -0.335 | -0.2651 | Yes |
| 220 | PMAIP1 | NA | 52731 | -0.336 | -0.2584 | Yes |
| 221 | MX2 | NA | 52778 | -0.337 | -0.2523 | Yes |
| 222 | STAT1 | NA | 52795 | -0.338 | -0.2455 | Yes |
| 223 | TRIM5 | NA | 52876 | -0.341 | -0.2399 | Yes |
| 224 | IRF1 | NA | 52959 | -0.344 | -0.2343 | Yes |
| 225 | RIOK3 | NA | 52987 | -0.346 | -0.2276 | Yes |
| 226 | TBK1 | NA | 53174 | -0.353 | -0.2237 | Yes |
| 227 | IFI27 | NA | 53187 | -0.354 | -0.2165 | Yes |
| 228 | PARP9 | NA | 53342 | -0.363 | -0.2118 | Yes |
| 229 | EXOC1 | NA | 53417 | -0.366 | -0.2056 | Yes |
| 230 | AIM2 | NA | 53450 | -0.368 | -0.1985 | Yes |
| 231 | CRCP | NA | 53451 | -0.368 | -0.1909 | Yes |
| 232 | APOBEC3F | NA | 53628 | -0.379 | -0.1862 | Yes |
| 233 | STAT2 | NA | 53669 | -0.382 | -0.1790 | Yes |
| 234 | IFIH1 | NA | 53720 | -0.385 | -0.1719 | Yes |
| 235 | RNASEL | NA | 53901 | -0.400 | -0.1669 | Yes |
| 236 | DDX60 | NA | 53952 | -0.404 | -0.1595 | Yes |
| 237 | IFIT2 | NA | 53965 | -0.406 | -0.1512 | Yes |
| 238 | CXCL9 | NA | 53980 | -0.407 | -0.1430 | Yes |
| 239 | IFNA13 | NA | 54077 | -0.417 | -0.1361 | Yes |
| 240 | PML | NA | 54084 | -0.419 | -0.1276 | Yes |
| 241 | IRF3 | NA | 54095 | -0.420 | -0.1190 | Yes |
| 242 | IL33 | NA | 54180 | -0.432 | -0.1116 | Yes |
| 243 | RTP4 | NA | 54182 | -0.432 | -0.1026 | Yes |
| 244 | APOBEC3C | NA | 54239 | -0.439 | -0.0945 | Yes |
| 245 | CD40 | NA | 54257 | -0.441 | -0.0857 | Yes |
| 246 | IL15 | NA | 54287 | -0.444 | -0.0770 | Yes |
| 247 | APOBEC3G | NA | 54303 | -0.448 | -0.0679 | Yes |
| 248 | DDX58 | NA | 54348 | -0.459 | -0.0592 | Yes |
| 249 | TLR3 | NA | 54368 | -0.464 | -0.0499 | Yes |
| 250 | APOBEC3D | NA | 54391 | -0.470 | -0.0406 | Yes |
| 251 | GBP1 | NA | 54416 | -0.475 | -0.0311 | Yes |
| 252 | TRIM22 | NA | 54536 | -0.540 | -0.0221 | Yes |
| 253 | MICA | NA | 54547 | -0.553 | -0.0108 | Yes |
| 254 | GBP3 | NA | 54553 | -0.558 | 0.0007 | Yes |