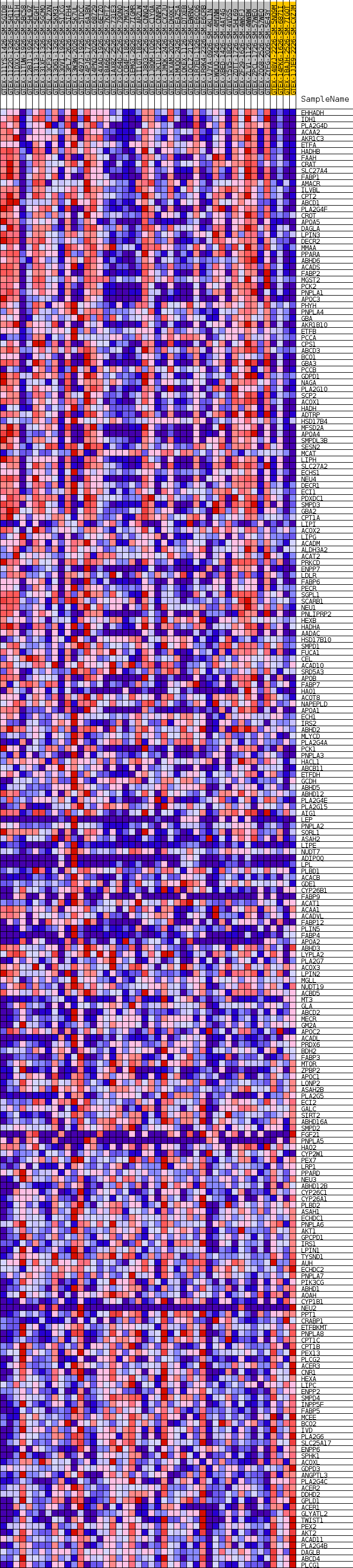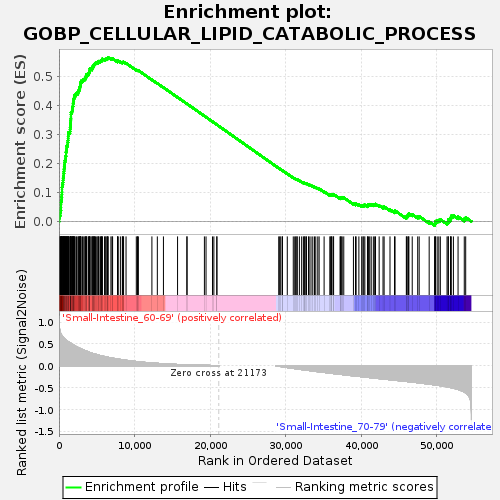
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Small-Intestine.Small-Intestine.cls #Small-Intestine_60-69_versus_Small-Intestine_70-79.Small-Intestine.cls #Small-Intestine_60-69_versus_Small-Intestine_70-79_repos |
| Phenotype | Small-Intestine.cls#Small-Intestine_60-69_versus_Small-Intestine_70-79_repos |
| Upregulated in class | Small-Intestine_60-69 |
| GeneSet | GOBP_CELLULAR_LIPID_CATABOLIC_PROCESS |
| Enrichment Score (ES) | 0.56712747 |
| Normalized Enrichment Score (NES) | 1.3912766 |
| Nominal p-value | 0.10891089 |
| FDR q-value | 0.6928571 |
| FWER p-Value | 0.981 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | EHHADH | NA | 32 | 0.916 | 0.0114 | Yes |
| 2 | IDH1 | NA | 78 | 0.846 | 0.0217 | Yes |
| 3 | PLA2G4D | NA | 182 | 0.761 | 0.0298 | Yes |
| 4 | ACAA2 | NA | 221 | 0.739 | 0.0388 | Yes |
| 5 | AKR1C3 | NA | 254 | 0.729 | 0.0478 | Yes |
| 6 | ETFA | NA | 259 | 0.728 | 0.0573 | Yes |
| 7 | HADHB | NA | 264 | 0.726 | 0.0668 | Yes |
| 8 | FAAH | NA | 315 | 0.707 | 0.0751 | Yes |
| 9 | CRAT | NA | 335 | 0.700 | 0.0840 | Yes |
| 10 | SLC27A4 | NA | 375 | 0.689 | 0.0923 | Yes |
| 11 | FABP1 | NA | 385 | 0.688 | 0.1012 | Yes |
| 12 | AMACR | NA | 395 | 0.685 | 0.1100 | Yes |
| 13 | ILVBL | NA | 398 | 0.683 | 0.1190 | Yes |
| 14 | CPT2 | NA | 435 | 0.675 | 0.1272 | Yes |
| 15 | ABCD1 | NA | 512 | 0.657 | 0.1344 | Yes |
| 16 | PLA2G4F | NA | 535 | 0.652 | 0.1426 | Yes |
| 17 | CROT | NA | 550 | 0.649 | 0.1508 | Yes |
| 18 | APOA5 | NA | 601 | 0.637 | 0.1583 | Yes |
| 19 | DAGLA | NA | 616 | 0.634 | 0.1663 | Yes |
| 20 | LPIN3 | NA | 653 | 0.628 | 0.1739 | Yes |
| 21 | DECR2 | NA | 669 | 0.624 | 0.1819 | Yes |
| 22 | MMAA | NA | 693 | 0.621 | 0.1896 | Yes |
| 23 | PPARA | NA | 730 | 0.614 | 0.1970 | Yes |
| 24 | ABHD6 | NA | 755 | 0.611 | 0.2046 | Yes |
| 25 | ACADS | NA | 791 | 0.606 | 0.2119 | Yes |
| 26 | FABP2 | NA | 866 | 0.596 | 0.2184 | Yes |
| 27 | MGST2 | NA | 875 | 0.594 | 0.2260 | Yes |
| 28 | PCK2 | NA | 927 | 0.587 | 0.2328 | Yes |
| 29 | PNPLA1 | NA | 928 | 0.587 | 0.2405 | Yes |
| 30 | APOC3 | NA | 1004 | 0.576 | 0.2467 | Yes |
| 31 | PHYH | NA | 1020 | 0.574 | 0.2540 | Yes |
| 32 | PNPLA4 | NA | 1028 | 0.573 | 0.2614 | Yes |
| 33 | GBA | NA | 1142 | 0.558 | 0.2666 | Yes |
| 34 | AKR1B10 | NA | 1151 | 0.556 | 0.2738 | Yes |
| 35 | ETFB | NA | 1188 | 0.552 | 0.2804 | Yes |
| 36 | PCCA | NA | 1208 | 0.550 | 0.2872 | Yes |
| 37 | CPS1 | NA | 1215 | 0.550 | 0.2943 | Yes |
| 38 | ABCD3 | NA | 1230 | 0.549 | 0.3013 | Yes |
| 39 | BCO1 | NA | 1247 | 0.547 | 0.3082 | Yes |
| 40 | GBA3 | NA | 1443 | 0.525 | 0.3115 | Yes |
| 41 | PCCB | NA | 1466 | 0.522 | 0.3179 | Yes |
| 42 | GDPD1 | NA | 1478 | 0.522 | 0.3246 | Yes |
| 43 | NAGA | NA | 1512 | 0.518 | 0.3308 | Yes |
| 44 | PLA2G10 | NA | 1531 | 0.516 | 0.3372 | Yes |
| 45 | SCP2 | NA | 1544 | 0.515 | 0.3438 | Yes |
| 46 | ACOX1 | NA | 1554 | 0.514 | 0.3504 | Yes |
| 47 | HADH | NA | 1572 | 0.512 | 0.3568 | Yes |
| 48 | ADTRP | NA | 1582 | 0.510 | 0.3633 | Yes |
| 49 | HSD17B4 | NA | 1584 | 0.510 | 0.3700 | Yes |
| 50 | MFSD2A | NA | 1592 | 0.510 | 0.3766 | Yes |
| 51 | APOA4 | NA | 1707 | 0.497 | 0.3810 | Yes |
| 52 | SMPDL3B | NA | 1759 | 0.490 | 0.3865 | Yes |
| 53 | SESN2 | NA | 1796 | 0.486 | 0.3922 | Yes |
| 54 | MCAT | NA | 1817 | 0.484 | 0.3982 | Yes |
| 55 | LIPH | NA | 1866 | 0.480 | 0.4036 | Yes |
| 56 | SLC27A2 | NA | 1870 | 0.479 | 0.4099 | Yes |
| 57 | ECHS1 | NA | 1940 | 0.470 | 0.4148 | Yes |
| 58 | NEU4 | NA | 1951 | 0.469 | 0.4208 | Yes |
| 59 | DECR1 | NA | 1977 | 0.466 | 0.4264 | Yes |
| 60 | ECI1 | NA | 2028 | 0.461 | 0.4316 | Yes |
| 61 | PDXDC1 | NA | 2063 | 0.458 | 0.4370 | Yes |
| 62 | SMPD3 | NA | 2233 | 0.442 | 0.4397 | Yes |
| 63 | GBA2 | NA | 2309 | 0.435 | 0.4440 | Yes |
| 64 | CPT1A | NA | 2509 | 0.417 | 0.4458 | Yes |
| 65 | LIPI | NA | 2549 | 0.414 | 0.4505 | Yes |
| 66 | ACOX2 | NA | 2623 | 0.408 | 0.4545 | Yes |
| 67 | LIPG | NA | 2723 | 0.400 | 0.4580 | Yes |
| 68 | ACADM | NA | 2758 | 0.397 | 0.4626 | Yes |
| 69 | ALDH3A2 | NA | 2774 | 0.396 | 0.4675 | Yes |
| 70 | ACAT2 | NA | 2829 | 0.391 | 0.4716 | Yes |
| 71 | PRKCD | NA | 2847 | 0.390 | 0.4764 | Yes |
| 72 | ENPP7 | NA | 2880 | 0.387 | 0.4809 | Yes |
| 73 | LDLR | NA | 2917 | 0.385 | 0.4853 | Yes |
| 74 | FABP6 | NA | 3104 | 0.370 | 0.4868 | Yes |
| 75 | PECR | NA | 3190 | 0.363 | 0.4900 | Yes |
| 76 | SGPL1 | NA | 3332 | 0.354 | 0.4920 | Yes |
| 77 | SCARB1 | NA | 3468 | 0.345 | 0.4941 | Yes |
| 78 | NEU1 | NA | 3502 | 0.343 | 0.4980 | Yes |
| 79 | PNLIPRP2 | NA | 3592 | 0.337 | 0.5008 | Yes |
| 80 | HEXB | NA | 3596 | 0.336 | 0.5051 | Yes |
| 81 | HADHA | NA | 3632 | 0.333 | 0.5089 | Yes |
| 82 | AADAC | NA | 3867 | 0.316 | 0.5087 | Yes |
| 83 | HSD17B10 | NA | 3888 | 0.315 | 0.5125 | Yes |
| 84 | SMPD1 | NA | 3989 | 0.309 | 0.5147 | Yes |
| 85 | FUCA1 | NA | 4033 | 0.306 | 0.5179 | Yes |
| 86 | CEL | NA | 4070 | 0.304 | 0.5213 | Yes |
| 87 | ACAD10 | NA | 4090 | 0.303 | 0.5249 | Yes |
| 88 | SRD5A3 | NA | 4098 | 0.303 | 0.5288 | Yes |
| 89 | APOB | NA | 4372 | 0.286 | 0.5275 | Yes |
| 90 | FABP7 | NA | 4385 | 0.286 | 0.5310 | Yes |
| 91 | HAO1 | NA | 4401 | 0.285 | 0.5345 | Yes |
| 92 | ACOT8 | NA | 4535 | 0.278 | 0.5357 | Yes |
| 93 | NAPEPLD | NA | 4603 | 0.275 | 0.5381 | Yes |
| 94 | APOA1 | NA | 4608 | 0.274 | 0.5416 | Yes |
| 95 | ECH1 | NA | 4695 | 0.270 | 0.5436 | Yes |
| 96 | IRS2 | NA | 4744 | 0.268 | 0.5462 | Yes |
| 97 | ABHD2 | NA | 4827 | 0.263 | 0.5482 | Yes |
| 98 | MLYCD | NA | 4993 | 0.255 | 0.5485 | Yes |
| 99 | PLA2G4A | NA | 5160 | 0.247 | 0.5487 | Yes |
| 100 | PCK1 | NA | 5170 | 0.246 | 0.5517 | Yes |
| 101 | PNPLA3 | NA | 5223 | 0.244 | 0.5540 | Yes |
| 102 | HACL1 | NA | 5423 | 0.234 | 0.5534 | Yes |
| 103 | ABCB11 | NA | 5485 | 0.231 | 0.5553 | Yes |
| 104 | ETFDH | NA | 5625 | 0.225 | 0.5557 | Yes |
| 105 | GCDH | NA | 5665 | 0.224 | 0.5579 | Yes |
| 106 | ABHD5 | NA | 5691 | 0.223 | 0.5604 | Yes |
| 107 | ABHD12 | NA | 5740 | 0.221 | 0.5624 | Yes |
| 108 | PLA2G4E | NA | 6031 | 0.209 | 0.5598 | Yes |
| 109 | PLA2G15 | NA | 6117 | 0.206 | 0.5610 | Yes |
| 110 | AIG1 | NA | 6225 | 0.203 | 0.5617 | Yes |
| 111 | LEP | NA | 6364 | 0.198 | 0.5617 | Yes |
| 112 | PNPLA2 | NA | 6406 | 0.196 | 0.5636 | Yes |
| 113 | SORL1 | NA | 6476 | 0.194 | 0.5648 | Yes |
| 114 | ASAH2 | NA | 6491 | 0.193 | 0.5671 | Yes |
| 115 | LIPE | NA | 6873 | 0.180 | 0.5625 | No |
| 116 | NUDT7 | NA | 7023 | 0.175 | 0.5620 | No |
| 117 | ADIPOQ | NA | 7095 | 0.173 | 0.5630 | No |
| 118 | LPL | NA | 7744 | 0.154 | 0.5531 | No |
| 119 | PLBD1 | NA | 7765 | 0.153 | 0.5548 | No |
| 120 | ACACB | NA | 7836 | 0.151 | 0.5555 | No |
| 121 | GDE1 | NA | 8125 | 0.144 | 0.5520 | No |
| 122 | CYP26B1 | NA | 8382 | 0.137 | 0.5491 | No |
| 123 | FABP9 | NA | 8416 | 0.136 | 0.5503 | No |
| 124 | ACAT1 | NA | 8486 | 0.135 | 0.5508 | No |
| 125 | ACAA1 | NA | 8547 | 0.133 | 0.5515 | No |
| 126 | ACADVL | NA | 8875 | 0.125 | 0.5471 | No |
| 127 | FABP12 | NA | 10233 | 0.096 | 0.5234 | No |
| 128 | PLIN5 | NA | 10371 | 0.094 | 0.5221 | No |
| 129 | FABP4 | NA | 10405 | 0.093 | 0.5227 | No |
| 130 | APOA2 | NA | 10506 | 0.091 | 0.5221 | No |
| 131 | ABHD3 | NA | 12298 | 0.062 | 0.4899 | No |
| 132 | LYPLA2 | NA | 13017 | 0.054 | 0.4774 | No |
| 133 | PLA2G7 | NA | 13825 | 0.045 | 0.4632 | No |
| 134 | ACOX3 | NA | 15695 | 0.029 | 0.4292 | No |
| 135 | LPIN2 | NA | 16927 | 0.021 | 0.4068 | No |
| 136 | MGLL | NA | 16952 | 0.021 | 0.4066 | No |
| 137 | NUDT19 | NA | 19248 | 0.009 | 0.3646 | No |
| 138 | ACBD5 | NA | 19463 | 0.008 | 0.3607 | No |
| 139 | MT3 | NA | 20319 | 0.005 | 0.3451 | No |
| 140 | GLA | NA | 20501 | 0.005 | 0.3418 | No |
| 141 | ABCD2 | NA | 20894 | 0.003 | 0.3346 | No |
| 142 | MECR | NA | 20901 | 0.003 | 0.3346 | No |
| 143 | GM2A | NA | 29087 | -0.010 | 0.1841 | No |
| 144 | APOC2 | NA | 29212 | -0.013 | 0.1820 | No |
| 145 | ACADL | NA | 29353 | -0.018 | 0.1797 | No |
| 146 | PRDX6 | NA | 29550 | -0.022 | 0.1764 | No |
| 147 | BDH2 | NA | 29585 | -0.023 | 0.1760 | No |
| 148 | FABP3 | NA | 30219 | -0.041 | 0.1649 | No |
| 149 | MTOR | NA | 31013 | -0.060 | 0.1511 | No |
| 150 | ZPBP2 | NA | 31208 | -0.064 | 0.1484 | No |
| 151 | APOC1 | NA | 31328 | -0.067 | 0.1471 | No |
| 152 | LONP2 | NA | 31481 | -0.072 | 0.1453 | No |
| 153 | ASAH2B | NA | 31540 | -0.073 | 0.1452 | No |
| 154 | PLA2G5 | NA | 31821 | -0.080 | 0.1411 | No |
| 155 | ECI2 | NA | 32124 | -0.087 | 0.1366 | No |
| 156 | GALC | NA | 32351 | -0.091 | 0.1337 | No |
| 157 | SIRT2 | NA | 32466 | -0.094 | 0.1328 | No |
| 158 | ABHD16A | NA | 32540 | -0.096 | 0.1327 | No |
| 159 | SMPD2 | NA | 32682 | -0.099 | 0.1314 | No |
| 160 | FGF21 | NA | 32767 | -0.101 | 0.1312 | No |
| 161 | PNPLA5 | NA | 33033 | -0.107 | 0.1278 | No |
| 162 | HAO2 | NA | 33111 | -0.109 | 0.1278 | No |
| 163 | CYP2W1 | NA | 33358 | -0.114 | 0.1248 | No |
| 164 | PEX7 | NA | 33595 | -0.119 | 0.1220 | No |
| 165 | LRP1 | NA | 33820 | -0.125 | 0.1195 | No |
| 166 | PPARD | NA | 33993 | -0.129 | 0.1180 | No |
| 167 | NEU3 | NA | 34259 | -0.135 | 0.1149 | No |
| 168 | ABHD12B | NA | 34434 | -0.138 | 0.1135 | No |
| 169 | CYP26C1 | NA | 35095 | -0.152 | 0.1034 | No |
| 170 | CYP26A1 | NA | 35852 | -0.168 | 0.0917 | No |
| 171 | PLBD2 | NA | 35958 | -0.171 | 0.0920 | No |
| 172 | ASAH1 | NA | 36085 | -0.173 | 0.0920 | No |
| 173 | ECHDC1 | NA | 36103 | -0.174 | 0.0939 | No |
| 174 | PNPLA6 | NA | 36292 | -0.178 | 0.0928 | No |
| 175 | AKT1 | NA | 36351 | -0.178 | 0.0941 | No |
| 176 | GPCPD1 | NA | 37207 | -0.195 | 0.0809 | No |
| 177 | IRS1 | NA | 37275 | -0.196 | 0.0823 | No |
| 178 | LPIN1 | NA | 37373 | -0.198 | 0.0831 | No |
| 179 | TYSND1 | NA | 37488 | -0.201 | 0.0836 | No |
| 180 | AUH | NA | 37697 | -0.206 | 0.0825 | No |
| 181 | ECHDC2 | NA | 38977 | -0.230 | 0.0620 | No |
| 182 | PNPLA7 | NA | 39250 | -0.235 | 0.0601 | No |
| 183 | PIK3CG | NA | 39332 | -0.237 | 0.0617 | No |
| 184 | ABHD1 | NA | 39697 | -0.243 | 0.0582 | No |
| 185 | AOAH | NA | 40069 | -0.251 | 0.0547 | No |
| 186 | CYP1B1 | NA | 40232 | -0.254 | 0.0550 | No |
| 187 | NEU2 | NA | 40344 | -0.256 | 0.0564 | No |
| 188 | PPT1 | NA | 40487 | -0.259 | 0.0572 | No |
| 189 | CRABP1 | NA | 40835 | -0.265 | 0.0543 | No |
| 190 | ETFBKMT | NA | 40873 | -0.266 | 0.0571 | No |
| 191 | PNPLA8 | NA | 40985 | -0.268 | 0.0586 | No |
| 192 | CPT1C | NA | 41133 | -0.271 | 0.0594 | No |
| 193 | CPT1B | NA | 41329 | -0.274 | 0.0594 | No |
| 194 | PEX13 | NA | 41612 | -0.279 | 0.0579 | No |
| 195 | PLCG2 | NA | 41794 | -0.282 | 0.0583 | No |
| 196 | ACER3 | NA | 41900 | -0.284 | 0.0601 | No |
| 197 | CNR1 | NA | 42381 | -0.293 | 0.0551 | No |
| 198 | HEXA | NA | 42894 | -0.302 | 0.0497 | No |
| 199 | LIPC | NA | 43038 | -0.305 | 0.0511 | No |
| 200 | ENPP2 | NA | 43817 | -0.319 | 0.0409 | No |
| 201 | SMPD4 | NA | 44416 | -0.330 | 0.0343 | No |
| 202 | INPP5F | NA | 44486 | -0.332 | 0.0374 | No |
| 203 | FABP5 | NA | 45979 | -0.359 | 0.0146 | No |
| 204 | MCEE | NA | 46023 | -0.360 | 0.0186 | No |
| 205 | BCO2 | NA | 46176 | -0.363 | 0.0205 | No |
| 206 | IVD | NA | 46280 | -0.365 | 0.0234 | No |
| 207 | PLA2G6 | NA | 46310 | -0.365 | 0.0277 | No |
| 208 | SLC25A17 | NA | 46760 | -0.373 | 0.0243 | No |
| 209 | ENPP6 | NA | 47486 | -0.388 | 0.0161 | No |
| 210 | SPHK1 | NA | 47680 | -0.391 | 0.0177 | No |
| 211 | ACOXL | NA | 49003 | -0.420 | -0.0011 | No |
| 212 | GDPD3 | NA | 49718 | -0.436 | -0.0085 | No |
| 213 | ANGPTL3 | NA | 49803 | -0.439 | -0.0043 | No |
| 214 | PLA2G4C | NA | 49830 | -0.440 | 0.0010 | No |
| 215 | ACER2 | NA | 50062 | -0.445 | 0.0026 | No |
| 216 | DDHD2 | NA | 50281 | -0.450 | 0.0045 | No |
| 217 | GPLD1 | NA | 50481 | -0.455 | 0.0068 | No |
| 218 | ACER1 | NA | 51369 | -0.481 | -0.0032 | No |
| 219 | GLYATL2 | NA | 51524 | -0.486 | 0.0004 | No |
| 220 | TWIST1 | NA | 51534 | -0.487 | 0.0066 | No |
| 221 | PEX2 | NA | 51828 | -0.497 | 0.0077 | No |
| 222 | AKT2 | NA | 51869 | -0.499 | 0.0136 | No |
| 223 | ACAD11 | NA | 51920 | -0.501 | 0.0192 | No |
| 224 | PLA2G4B | NA | 52210 | -0.512 | 0.0206 | No |
| 225 | DAGLB | NA | 52827 | -0.541 | 0.0164 | No |
| 226 | ABCD4 | NA | 53650 | -0.599 | 0.0092 | No |
| 227 | PLCG1 | NA | 53838 | -0.619 | 0.0139 | No |