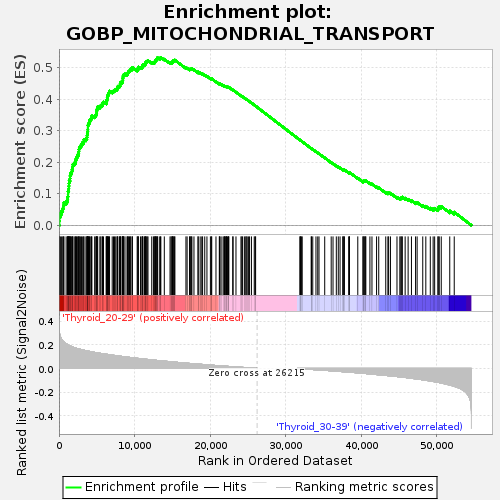
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Thyroid.Thyroid.cls #Thyroid_20-29_versus_Thyroid_30-39.Thyroid.cls #Thyroid_20-29_versus_Thyroid_30-39_repos |
| Phenotype | Thyroid.cls#Thyroid_20-29_versus_Thyroid_30-39_repos |
| Upregulated in class | Thyroid_20-29 |
| GeneSet | GOBP_MITOCHONDRIAL_TRANSPORT |
| Enrichment Score (ES) | 0.53227144 |
| Normalized Enrichment Score (NES) | 1.6131521 |
| Nominal p-value | 0.014198783 |
| FDR q-value | 0.5042422 |
| FWER p-Value | 0.843 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | BMF | NA | 51 | 0.308 | 0.0133 | Yes |
| 2 | TMEM102 | NA | 62 | 0.301 | 0.0271 | Yes |
| 3 | YWHAE | NA | 183 | 0.269 | 0.0373 | Yes |
| 4 | TIMM8B | NA | 346 | 0.244 | 0.0457 | Yes |
| 5 | ACAA2 | NA | 497 | 0.231 | 0.0536 | Yes |
| 6 | SLC25A52 | NA | 580 | 0.227 | 0.0626 | Yes |
| 7 | CNP | NA | 656 | 0.223 | 0.0716 | Yes |
| 8 | NDUFA13 | NA | 946 | 0.208 | 0.0759 | Yes |
| 9 | UBL5 | NA | 1120 | 0.201 | 0.0820 | Yes |
| 10 | TIMM17B | NA | 1161 | 0.200 | 0.0905 | Yes |
| 11 | SFN | NA | 1197 | 0.199 | 0.0991 | Yes |
| 12 | SFXN5 | NA | 1200 | 0.199 | 0.1082 | Yes |
| 13 | TIMM17A | NA | 1292 | 0.195 | 0.1156 | Yes |
| 14 | YWHAH | NA | 1304 | 0.195 | 0.1244 | Yes |
| 15 | SAE1 | NA | 1323 | 0.194 | 0.1331 | Yes |
| 16 | TIMM23 | NA | 1345 | 0.194 | 0.1417 | Yes |
| 17 | BCL2 | NA | 1456 | 0.190 | 0.1485 | Yes |
| 18 | ROMO1 | NA | 1468 | 0.190 | 0.1570 | Yes |
| 19 | SIRT4 | NA | 1507 | 0.189 | 0.1651 | Yes |
| 20 | MIPEP | NA | 1672 | 0.184 | 0.1706 | Yes |
| 21 | TOMM5 | NA | 1732 | 0.182 | 0.1779 | Yes |
| 22 | HSPA4 | NA | 1803 | 0.181 | 0.1850 | Yes |
| 23 | YWHAZ | NA | 1825 | 0.180 | 0.1930 | Yes |
| 24 | MPC2 | NA | 2015 | 0.176 | 0.1976 | Yes |
| 25 | VDAC1 | NA | 2174 | 0.172 | 0.2027 | Yes |
| 26 | DNAJC15 | NA | 2218 | 0.172 | 0.2099 | Yes |
| 27 | MAIP1 | NA | 2320 | 0.170 | 0.2159 | Yes |
| 28 | SLC25A20 | NA | 2425 | 0.168 | 0.2217 | Yes |
| 29 | UBE2L3 | NA | 2547 | 0.166 | 0.2272 | Yes |
| 30 | TIMM22 | NA | 2620 | 0.164 | 0.2335 | Yes |
| 31 | SLC25A5 | NA | 2636 | 0.164 | 0.2408 | Yes |
| 32 | TOMM34 | NA | 2683 | 0.163 | 0.2475 | Yes |
| 33 | SLC25A4 | NA | 2833 | 0.161 | 0.2522 | Yes |
| 34 | NMT1 | NA | 2972 | 0.158 | 0.2570 | Yes |
| 35 | UBE2J2 | NA | 3088 | 0.157 | 0.2621 | Yes |
| 36 | TMEM14DP | NA | 3262 | 0.154 | 0.2661 | Yes |
| 37 | UCP2 | NA | 3306 | 0.153 | 0.2724 | Yes |
| 38 | DYNLT1 | NA | 3565 | 0.150 | 0.2746 | Yes |
| 39 | LMAN1 | NA | 3687 | 0.148 | 0.2792 | Yes |
| 40 | SREBF2 | NA | 3717 | 0.148 | 0.2855 | Yes |
| 41 | TIMM13 | NA | 3744 | 0.147 | 0.2918 | Yes |
| 42 | TMEM14B | NA | 3795 | 0.147 | 0.2977 | Yes |
| 43 | HSP90AA1 | NA | 3818 | 0.146 | 0.3041 | Yes |
| 44 | TOMM22 | NA | 3826 | 0.146 | 0.3107 | Yes |
| 45 | GRPEL2 | NA | 3827 | 0.146 | 0.3175 | Yes |
| 46 | SMDT1 | NA | 3904 | 0.145 | 0.3228 | Yes |
| 47 | PMPCA | NA | 4003 | 0.144 | 0.3277 | Yes |
| 48 | BCAP31 | NA | 4043 | 0.143 | 0.3336 | Yes |
| 49 | BAD | NA | 4205 | 0.141 | 0.3372 | Yes |
| 50 | TFDP2 | NA | 4302 | 0.140 | 0.3419 | Yes |
| 51 | TOMM40L | NA | 4355 | 0.140 | 0.3474 | Yes |
| 52 | TOMM40 | NA | 4721 | 0.135 | 0.3470 | Yes |
| 53 | TOMM70 | NA | 4871 | 0.133 | 0.3504 | Yes |
| 54 | TIMM50 | NA | 4959 | 0.132 | 0.3550 | Yes |
| 55 | MUL1 | NA | 4996 | 0.132 | 0.3604 | Yes |
| 56 | TMEM14C | NA | 5009 | 0.132 | 0.3663 | Yes |
| 57 | ATG13 | NA | 5058 | 0.131 | 0.3715 | Yes |
| 58 | PDCD5 | NA | 5122 | 0.131 | 0.3764 | Yes |
| 59 | SLC25A41 | NA | 5410 | 0.128 | 0.3770 | Yes |
| 60 | SLC25A2 | NA | 5531 | 0.126 | 0.3807 | Yes |
| 61 | FZD5 | NA | 5712 | 0.125 | 0.3831 | Yes |
| 62 | HSPD1 | NA | 5757 | 0.124 | 0.3881 | Yes |
| 63 | HEBP2 | NA | 5890 | 0.123 | 0.3913 | Yes |
| 64 | TIMM21 | NA | 6259 | 0.120 | 0.3901 | Yes |
| 65 | BAK1 | NA | 6262 | 0.120 | 0.3956 | Yes |
| 66 | MRPL18 | NA | 6354 | 0.119 | 0.3994 | Yes |
| 67 | GRPEL1 | NA | 6381 | 0.118 | 0.4044 | Yes |
| 68 | BAG4 | NA | 6408 | 0.118 | 0.4094 | Yes |
| 69 | LETM1 | NA | 6476 | 0.117 | 0.4136 | Yes |
| 70 | YWHAB | NA | 6551 | 0.117 | 0.4177 | Yes |
| 71 | YWHAQ | NA | 6621 | 0.116 | 0.4218 | Yes |
| 72 | AIFM1 | NA | 6691 | 0.115 | 0.4258 | Yes |
| 73 | TSPO | NA | 7042 | 0.112 | 0.4246 | Yes |
| 74 | SLC25A1 | NA | 7216 | 0.111 | 0.4265 | Yes |
| 75 | MTX2 | NA | 7351 | 0.109 | 0.4291 | Yes |
| 76 | STPG1 | NA | 7509 | 0.108 | 0.4313 | Yes |
| 77 | FBXO7 | NA | 7677 | 0.107 | 0.4331 | Yes |
| 78 | UCP3 | NA | 7737 | 0.106 | 0.4370 | Yes |
| 79 | STOML2 | NA | 7788 | 0.106 | 0.4409 | Yes |
| 80 | FIS1 | NA | 7998 | 0.104 | 0.4419 | Yes |
| 81 | TP63 | NA | 8080 | 0.103 | 0.4452 | Yes |
| 82 | UBE2D3 | NA | 8099 | 0.103 | 0.4496 | Yes |
| 83 | VPS35 | NA | 8136 | 0.103 | 0.4537 | Yes |
| 84 | PARL | NA | 8340 | 0.101 | 0.4547 | Yes |
| 85 | HUWE1 | NA | 8430 | 0.100 | 0.4577 | Yes |
| 86 | SLC9A1 | NA | 8451 | 0.100 | 0.4620 | Yes |
| 87 | PSEN1 | NA | 8454 | 0.100 | 0.4665 | Yes |
| 88 | SLC35F6 | NA | 8475 | 0.100 | 0.4708 | Yes |
| 89 | BCL2L1 | NA | 8495 | 0.100 | 0.4751 | Yes |
| 90 | CHCHD10 | NA | 8568 | 0.099 | 0.4783 | Yes |
| 91 | BAX | NA | 8734 | 0.098 | 0.4798 | Yes |
| 92 | MAPK8 | NA | 9002 | 0.096 | 0.4793 | Yes |
| 93 | CPT1A | NA | 9075 | 0.095 | 0.4824 | Yes |
| 94 | NPEPPS | NA | 9136 | 0.095 | 0.4857 | Yes |
| 95 | PITRM1 | NA | 9218 | 0.094 | 0.4885 | Yes |
| 96 | TIMM44 | NA | 9335 | 0.093 | 0.4907 | Yes |
| 97 | VDAC2 | NA | 9433 | 0.092 | 0.4932 | Yes |
| 98 | PSEN2 | NA | 9512 | 0.092 | 0.4960 | Yes |
| 99 | PRKAA2 | NA | 9700 | 0.090 | 0.4968 | Yes |
| 100 | SFXN1 | NA | 9708 | 0.090 | 0.5008 | Yes |
| 101 | SH3GLB1 | NA | 10348 | 0.085 | 0.4930 | Yes |
| 102 | PPP1R13B | NA | 10447 | 0.085 | 0.4951 | Yes |
| 103 | MPC1L | NA | 10455 | 0.085 | 0.4989 | Yes |
| 104 | CHCHD4 | NA | 10507 | 0.084 | 0.5019 | Yes |
| 105 | ALKBH7 | NA | 10785 | 0.082 | 0.5006 | Yes |
| 106 | HSPA1L | NA | 10975 | 0.081 | 0.5009 | Yes |
| 107 | GSK3B | NA | 11040 | 0.080 | 0.5034 | Yes |
| 108 | MCUR1 | NA | 11043 | 0.080 | 0.5071 | Yes |
| 109 | TST | NA | 11120 | 0.080 | 0.5094 | Yes |
| 110 | MFN2 | NA | 11324 | 0.078 | 0.5093 | Yes |
| 111 | BNIP3L | NA | 11405 | 0.078 | 0.5114 | Yes |
| 112 | CPT2 | NA | 11423 | 0.078 | 0.5147 | Yes |
| 113 | HAX1 | NA | 11472 | 0.077 | 0.5174 | Yes |
| 114 | BAP1 | NA | 11563 | 0.077 | 0.5193 | Yes |
| 115 | TMEM14EP | NA | 11677 | 0.076 | 0.5207 | Yes |
| 116 | TIMM10 | NA | 11780 | 0.075 | 0.5223 | Yes |
| 117 | YWHAG | NA | 12277 | 0.072 | 0.5165 | Yes |
| 118 | GSK3A | NA | 12499 | 0.070 | 0.5157 | Yes |
| 119 | ABCB10 | NA | 12603 | 0.070 | 0.5171 | Yes |
| 120 | PPP3R1 | NA | 12709 | 0.069 | 0.5183 | Yes |
| 121 | PINK1 | NA | 12722 | 0.069 | 0.5213 | Yes |
| 122 | TIMM10B | NA | 12757 | 0.069 | 0.5239 | Yes |
| 123 | OPA1 | NA | 12853 | 0.068 | 0.5253 | Yes |
| 124 | MICALL2 | NA | 12951 | 0.068 | 0.5266 | Yes |
| 125 | SLC39A8 | NA | 12989 | 0.067 | 0.5291 | Yes |
| 126 | SLC25A22 | NA | 13023 | 0.067 | 0.5316 | Yes |
| 127 | SLC25A23 | NA | 13277 | 0.066 | 0.5300 | Yes |
| 128 | SAMM50 | NA | 13443 | 0.065 | 0.5299 | Yes |
| 129 | ATF2 | NA | 13478 | 0.064 | 0.5323 | Yes |
| 130 | TOMM7 | NA | 13940 | 0.062 | 0.5266 | No |
| 131 | PNPT1 | NA | 14699 | 0.057 | 0.5153 | No |
| 132 | IMMP1L | NA | 14837 | 0.056 | 0.5154 | No |
| 133 | MIR29A | NA | 14947 | 0.055 | 0.5160 | No |
| 134 | DNAJC19 | NA | 14999 | 0.055 | 0.5176 | No |
| 135 | MOAP1 | NA | 15024 | 0.055 | 0.5197 | No |
| 136 | FLVCR1 | NA | 15114 | 0.054 | 0.5205 | No |
| 137 | ACACA | NA | 15123 | 0.054 | 0.5229 | No |
| 138 | MPV17L | NA | 15276 | 0.053 | 0.5226 | No |
| 139 | PAM16 | NA | 15343 | 0.053 | 0.5238 | No |
| 140 | BNIP3 | NA | 16804 | 0.045 | 0.4990 | No |
| 141 | SLC25A28 | NA | 16959 | 0.044 | 0.4982 | No |
| 142 | HK2 | NA | 17279 | 0.042 | 0.4943 | No |
| 143 | MICU1 | NA | 17340 | 0.042 | 0.4951 | No |
| 144 | PPP3CC | NA | 17438 | 0.041 | 0.4952 | No |
| 145 | SLC25A21 | NA | 17517 | 0.041 | 0.4957 | No |
| 146 | BLOC1S2 | NA | 17593 | 0.041 | 0.4962 | No |
| 147 | PPIF | NA | 17866 | 0.039 | 0.4930 | No |
| 148 | TP53 | NA | 18414 | 0.036 | 0.4846 | No |
| 149 | RHOU | NA | 18415 | 0.036 | 0.4863 | No |
| 150 | STARD3 | NA | 18687 | 0.035 | 0.4829 | No |
| 151 | TIMM9 | NA | 18902 | 0.034 | 0.4806 | No |
| 152 | HSPA1A | NA | 19009 | 0.033 | 0.4802 | No |
| 153 | BOK | NA | 19293 | 0.032 | 0.4765 | No |
| 154 | HTRA2 | NA | 19584 | 0.031 | 0.4725 | No |
| 155 | MFF | NA | 20041 | 0.028 | 0.4654 | No |
| 156 | SLC25A51 | NA | 20162 | 0.028 | 0.4645 | No |
| 157 | SLC25A13 | NA | 20218 | 0.027 | 0.4648 | No |
| 158 | TP53BP2 | NA | 20780 | 0.025 | 0.4556 | No |
| 159 | BBC3 | NA | 21231 | 0.022 | 0.4483 | No |
| 160 | SLC25A33 | NA | 21236 | 0.022 | 0.4493 | No |
| 161 | AGK | NA | 21332 | 0.022 | 0.4486 | No |
| 162 | AIP | NA | 21567 | 0.021 | 0.4452 | No |
| 163 | SPG7 | NA | 21813 | 0.020 | 0.4416 | No |
| 164 | AFG3L2 | NA | 21887 | 0.019 | 0.4412 | No |
| 165 | CSNK2A2 | NA | 21917 | 0.019 | 0.4415 | No |
| 166 | RHOT1 | NA | 22020 | 0.019 | 0.4405 | No |
| 167 | SLC25A37 | NA | 22109 | 0.018 | 0.4398 | No |
| 168 | IMMP2L | NA | 22169 | 0.018 | 0.4395 | No |
| 169 | TMEM14A | NA | 22174 | 0.018 | 0.4403 | No |
| 170 | ARIH2 | NA | 22312 | 0.017 | 0.4386 | No |
| 171 | MICU2 | NA | 22410 | 0.017 | 0.4376 | No |
| 172 | CASP8 | NA | 22431 | 0.017 | 0.4380 | No |
| 173 | CAMK2A | NA | 22487 | 0.017 | 0.4377 | No |
| 174 | MRS2 | NA | 23000 | 0.014 | 0.4290 | No |
| 175 | PRKAG2 | NA | 23044 | 0.014 | 0.4288 | No |
| 176 | SLC25A24 | NA | 23428 | 0.012 | 0.4223 | No |
| 177 | SFXN3 | NA | 24088 | 0.010 | 0.4107 | No |
| 178 | NOL3 | NA | 24199 | 0.009 | 0.4091 | No |
| 179 | RNF31 | NA | 24303 | 0.009 | 0.4076 | No |
| 180 | MCUB | NA | 24542 | 0.008 | 0.4036 | No |
| 181 | TIMM29 | NA | 24737 | 0.007 | 0.4003 | No |
| 182 | COX18 | NA | 24774 | 0.007 | 0.4000 | No |
| 183 | SLC8B1 | NA | 24913 | 0.006 | 0.3977 | No |
| 184 | MTRNR2L5 | NA | 25099 | 0.005 | 0.3945 | No |
| 185 | DNAJC30 | NA | 25227 | 0.005 | 0.3924 | No |
| 186 | TOMM20 | NA | 25475 | 0.004 | 0.3881 | No |
| 187 | SLC25A10 | NA | 25846 | 0.002 | 0.3814 | No |
| 188 | NAIF1 | NA | 25965 | 0.001 | 0.3792 | No |
| 189 | HIP1R | NA | 26028 | 0.001 | 0.3781 | No |
| 190 | BCS1L | NA | 31900 | -0.002 | 0.2702 | No |
| 191 | TFDP1 | NA | 31923 | -0.002 | 0.2699 | No |
| 192 | LEPROT | NA | 31975 | -0.002 | 0.2691 | No |
| 193 | TP73 | NA | 32091 | -0.003 | 0.2671 | No |
| 194 | VPS11 | NA | 32195 | -0.003 | 0.2653 | No |
| 195 | FZD9 | NA | 33382 | -0.008 | 0.2438 | No |
| 196 | SLC25A31 | NA | 33428 | -0.008 | 0.2434 | No |
| 197 | MTERF4 | NA | 33593 | -0.008 | 0.2408 | No |
| 198 | OXA1L | NA | 34016 | -0.010 | 0.2335 | No |
| 199 | SLC25A6 | NA | 34253 | -0.011 | 0.2296 | No |
| 200 | PRKAA1 | NA | 34419 | -0.012 | 0.2271 | No |
| 201 | SLC25A16 | NA | 35175 | -0.015 | 0.2139 | No |
| 202 | SLC25A32 | NA | 36033 | -0.019 | 0.1991 | No |
| 203 | PMPCB | NA | 36275 | -0.020 | 0.1956 | No |
| 204 | MCU | NA | 36710 | -0.022 | 0.1886 | No |
| 205 | DNLZ | NA | 36971 | -0.023 | 0.1849 | No |
| 206 | SLC25A38 | NA | 37205 | -0.025 | 0.1818 | No |
| 207 | ZNF205 | NA | 37562 | -0.026 | 0.1764 | No |
| 208 | SFXN2 | NA | 37621 | -0.027 | 0.1766 | No |
| 209 | SLC25A15 | NA | 37707 | -0.027 | 0.1763 | No |
| 210 | PRKAB2 | NA | 37784 | -0.027 | 0.1761 | No |
| 211 | EYA2 | NA | 38348 | -0.030 | 0.1672 | No |
| 212 | ACACB | NA | 38433 | -0.030 | 0.1670 | No |
| 213 | MT-ATP6 | NA | 38455 | -0.031 | 0.1681 | No |
| 214 | MID1IP1 | NA | 39553 | -0.036 | 0.1496 | No |
| 215 | FBXW7 | NA | 40242 | -0.040 | 0.1388 | No |
| 216 | THEM4 | NA | 40262 | -0.040 | 0.1403 | No |
| 217 | MGARP | NA | 40367 | -0.041 | 0.1402 | No |
| 218 | RAC2 | NA | 40471 | -0.041 | 0.1403 | No |
| 219 | TOMM20L | NA | 40549 | -0.042 | 0.1408 | No |
| 220 | BID | NA | 40618 | -0.042 | 0.1415 | No |
| 221 | SLC25A18 | NA | 41158 | -0.045 | 0.1337 | No |
| 222 | BAG3 | NA | 41430 | -0.047 | 0.1309 | No |
| 223 | USP36 | NA | 42039 | -0.051 | 0.1220 | No |
| 224 | CPT1B | NA | 42326 | -0.053 | 0.1192 | No |
| 225 | SLC25A12 | NA | 43255 | -0.058 | 0.1048 | No |
| 226 | RHOT2 | NA | 43540 | -0.060 | 0.1024 | No |
| 227 | TIMM23B | NA | 43635 | -0.060 | 0.1034 | No |
| 228 | UBL4B | NA | 43877 | -0.062 | 0.1019 | No |
| 229 | LRRK2 | NA | 44741 | -0.068 | 0.0891 | No |
| 230 | UCP1 | NA | 45120 | -0.071 | 0.0855 | No |
| 231 | MPC1 | NA | 45250 | -0.072 | 0.0864 | No |
| 232 | SIAH3 | NA | 45367 | -0.072 | 0.0876 | No |
| 233 | E2F1 | NA | 45449 | -0.073 | 0.0895 | No |
| 234 | ABLIM3 | NA | 45823 | -0.075 | 0.0861 | No |
| 235 | SLC25A36 | NA | 46217 | -0.078 | 0.0825 | No |
| 236 | SFXN4 | NA | 46657 | -0.082 | 0.0782 | No |
| 237 | BCL2L11 | NA | 47203 | -0.087 | 0.0722 | No |
| 238 | HPS4 | NA | 47409 | -0.088 | 0.0725 | No |
| 239 | PMAIP1 | NA | 48153 | -0.095 | 0.0633 | No |
| 240 | SLC25A30 | NA | 48559 | -0.099 | 0.0604 | No |
| 241 | GDAP1 | NA | 49152 | -0.105 | 0.0544 | No |
| 242 | SREBF1 | NA | 49528 | -0.109 | 0.0525 | No |
| 243 | MT-ATP8 | NA | 49729 | -0.111 | 0.0540 | No |
| 244 | SLC25A29 | NA | 50160 | -0.116 | 0.0515 | No |
| 245 | SLC25A14 | NA | 50203 | -0.117 | 0.0561 | No |
| 246 | MICU3 | NA | 50340 | -0.119 | 0.0591 | No |
| 247 | C11orf65 | NA | 50602 | -0.122 | 0.0600 | No |
| 248 | GZMB | NA | 51728 | -0.139 | 0.0457 | No |
| 249 | THRSP | NA | 52321 | -0.150 | 0.0418 | No |