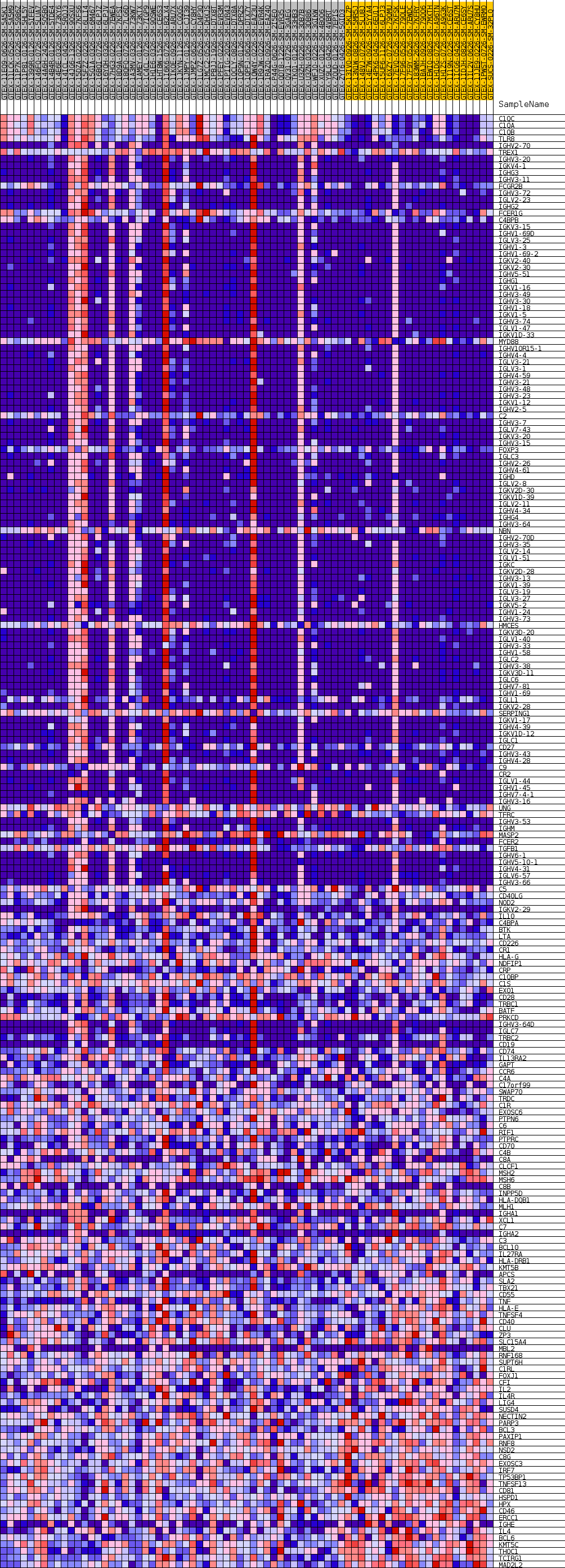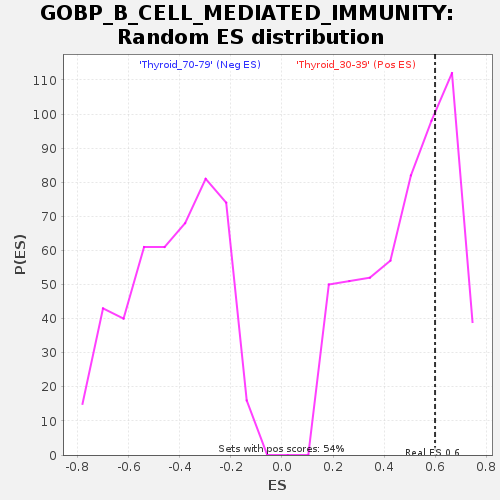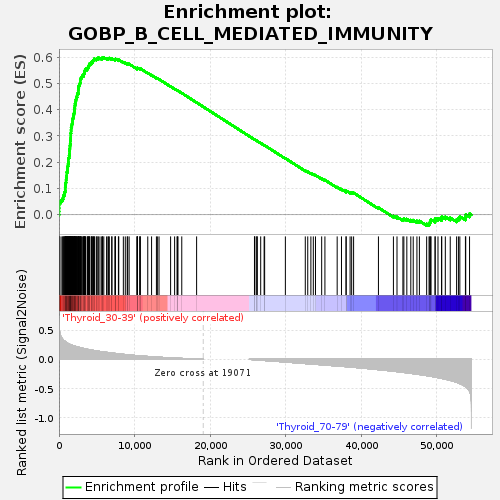
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Thyroid.Thyroid.cls #Thyroid_30-39_versus_Thyroid_70-79.Thyroid.cls #Thyroid_30-39_versus_Thyroid_70-79_repos |
| Phenotype | Thyroid.cls#Thyroid_30-39_versus_Thyroid_70-79_repos |
| Upregulated in class | Thyroid_30-39 |
| GeneSet | GOBP_B_CELL_MEDIATED_IMMUNITY |
| Enrichment Score (ES) | 0.5990685 |
| Normalized Enrichment Score (NES) | 1.2130545 |
| Nominal p-value | 0.3530499 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | C1QC | NA | 22 | 0.551 | 0.0125 | Yes |
| 2 | C1QA | NA | 28 | 0.541 | 0.0250 | Yes |
| 3 | C1QB | NA | 33 | 0.534 | 0.0374 | Yes |
| 4 | TLR8 | NA | 39 | 0.523 | 0.0496 | Yes |
| 5 | IGHV2-70 | NA | 266 | 0.384 | 0.0544 | Yes |
| 6 | TREX1 | NA | 437 | 0.350 | 0.0595 | Yes |
| 7 | IGHV3-20 | NA | 541 | 0.333 | 0.0654 | Yes |
| 8 | IGKV4-1 | NA | 570 | 0.329 | 0.0725 | Yes |
| 9 | IGHG3 | NA | 685 | 0.317 | 0.0779 | Yes |
| 10 | IGHV3-11 | NA | 702 | 0.315 | 0.0849 | Yes |
| 11 | FCGR2B | NA | 799 | 0.305 | 0.0903 | Yes |
| 12 | IGHV3-72 | NA | 821 | 0.303 | 0.0970 | Yes |
| 13 | IGLV2-23 | NA | 825 | 0.303 | 0.1040 | Yes |
| 14 | IGHG2 | NA | 846 | 0.301 | 0.1107 | Yes |
| 15 | FCER1G | NA | 868 | 0.298 | 0.1173 | Yes |
| 16 | C4BPB | NA | 906 | 0.294 | 0.1235 | Yes |
| 17 | IGKV3-15 | NA | 938 | 0.291 | 0.1297 | Yes |
| 18 | IGHV1-69D | NA | 947 | 0.290 | 0.1363 | Yes |
| 19 | IGLV3-25 | NA | 962 | 0.289 | 0.1428 | Yes |
| 20 | IGHV1-3 | NA | 1000 | 0.286 | 0.1488 | Yes |
| 21 | IGHV1-69-2 | NA | 1014 | 0.284 | 0.1552 | Yes |
| 22 | IGKV2-40 | NA | 1015 | 0.284 | 0.1619 | Yes |
| 23 | IGKV2-30 | NA | 1114 | 0.276 | 0.1665 | Yes |
| 24 | IGHV5-51 | NA | 1135 | 0.274 | 0.1726 | Yes |
| 25 | IGHG1 | NA | 1146 | 0.273 | 0.1788 | Yes |
| 26 | IGKV1-16 | NA | 1158 | 0.272 | 0.1849 | Yes |
| 27 | IGHV3-49 | NA | 1199 | 0.270 | 0.1905 | Yes |
| 28 | IGHV3-30 | NA | 1213 | 0.269 | 0.1965 | Yes |
| 29 | IGHV1-18 | NA | 1236 | 0.267 | 0.2024 | Yes |
| 30 | IGKV1-5 | NA | 1241 | 0.267 | 0.2086 | Yes |
| 31 | IGHV3-74 | NA | 1243 | 0.267 | 0.2148 | Yes |
| 32 | IGLV1-47 | NA | 1327 | 0.261 | 0.2193 | Yes |
| 33 | IGKV1D-33 | NA | 1341 | 0.260 | 0.2252 | Yes |
| 34 | MYD88 | NA | 1367 | 0.259 | 0.2308 | Yes |
| 35 | IGHV1OR15-1 | NA | 1378 | 0.258 | 0.2366 | Yes |
| 36 | IGHV4-4 | NA | 1379 | 0.258 | 0.2427 | Yes |
| 37 | IGLV3-21 | NA | 1420 | 0.255 | 0.2479 | Yes |
| 38 | IGLV3-1 | NA | 1435 | 0.255 | 0.2536 | Yes |
| 39 | IGHV4-59 | NA | 1451 | 0.254 | 0.2592 | Yes |
| 40 | IGHV3-21 | NA | 1460 | 0.253 | 0.2650 | Yes |
| 41 | IGHV3-48 | NA | 1478 | 0.252 | 0.2706 | Yes |
| 42 | IGHV3-23 | NA | 1510 | 0.251 | 0.2759 | Yes |
| 43 | IGKV1-12 | NA | 1511 | 0.251 | 0.2818 | Yes |
| 44 | IGHV2-5 | NA | 1536 | 0.249 | 0.2871 | Yes |
| 45 | C2 | NA | 1543 | 0.249 | 0.2928 | Yes |
| 46 | IGHV3-7 | NA | 1550 | 0.248 | 0.2985 | Yes |
| 47 | IGLV7-43 | NA | 1552 | 0.248 | 0.3043 | Yes |
| 48 | IGKV3-20 | NA | 1555 | 0.248 | 0.3101 | Yes |
| 49 | IGHV3-15 | NA | 1586 | 0.246 | 0.3153 | Yes |
| 50 | FOXP3 | NA | 1596 | 0.246 | 0.3209 | Yes |
| 51 | IGLC3 | NA | 1602 | 0.246 | 0.3265 | Yes |
| 52 | IGHV2-26 | NA | 1612 | 0.245 | 0.3321 | Yes |
| 53 | IGHV4-61 | NA | 1690 | 0.242 | 0.3363 | Yes |
| 54 | IGHD | NA | 1697 | 0.241 | 0.3419 | Yes |
| 55 | IGLV2-8 | NA | 1729 | 0.239 | 0.3469 | Yes |
| 56 | IGKV2D-30 | NA | 1768 | 0.238 | 0.3517 | Yes |
| 57 | IGKV1D-39 | NA | 1794 | 0.236 | 0.3568 | Yes |
| 58 | IGLV2-11 | NA | 1811 | 0.236 | 0.3620 | Yes |
| 59 | IGHV4-34 | NA | 1818 | 0.235 | 0.3674 | Yes |
| 60 | IGHG4 | NA | 1903 | 0.232 | 0.3713 | Yes |
| 61 | IGHV3-64 | NA | 1932 | 0.231 | 0.3761 | Yes |
| 62 | NBN | NA | 1962 | 0.230 | 0.3810 | Yes |
| 63 | IGHV2-70D | NA | 1979 | 0.229 | 0.3860 | Yes |
| 64 | IGHV3-35 | NA | 2011 | 0.228 | 0.3908 | Yes |
| 65 | IGLV2-14 | NA | 2033 | 0.227 | 0.3957 | Yes |
| 66 | IGLV1-51 | NA | 2044 | 0.227 | 0.4008 | Yes |
| 67 | IGKC | NA | 2057 | 0.226 | 0.4059 | Yes |
| 68 | IGKV2D-28 | NA | 2099 | 0.225 | 0.4104 | Yes |
| 69 | IGHV3-13 | NA | 2110 | 0.224 | 0.4155 | Yes |
| 70 | IGKV1-39 | NA | 2126 | 0.224 | 0.4204 | Yes |
| 71 | IGLV3-19 | NA | 2132 | 0.224 | 0.4256 | Yes |
| 72 | IGLV3-27 | NA | 2209 | 0.221 | 0.4293 | Yes |
| 73 | IGKV5-2 | NA | 2231 | 0.220 | 0.4341 | Yes |
| 74 | IGHV1-24 | NA | 2244 | 0.219 | 0.4390 | Yes |
| 75 | IGHV3-73 | NA | 2283 | 0.217 | 0.4434 | Yes |
| 76 | HMCES | NA | 2321 | 0.216 | 0.4477 | Yes |
| 77 | IGKV3D-20 | NA | 2398 | 0.213 | 0.4513 | Yes |
| 78 | IGLV1-40 | NA | 2400 | 0.213 | 0.4563 | Yes |
| 79 | IGHV3-33 | NA | 2419 | 0.212 | 0.4609 | Yes |
| 80 | IGHV1-58 | NA | 2539 | 0.208 | 0.4636 | Yes |
| 81 | IGLC2 | NA | 2547 | 0.208 | 0.4683 | Yes |
| 82 | IGHV3-38 | NA | 2548 | 0.208 | 0.4731 | Yes |
| 83 | IGKV3D-11 | NA | 2573 | 0.207 | 0.4775 | Yes |
| 84 | IGLC6 | NA | 2574 | 0.207 | 0.4824 | Yes |
| 85 | IGHV7-81 | NA | 2622 | 0.205 | 0.4863 | Yes |
| 86 | IGHV1-69 | NA | 2627 | 0.205 | 0.4910 | Yes |
| 87 | IGLL1 | NA | 2697 | 0.202 | 0.4945 | Yes |
| 88 | IGKV2-28 | NA | 2754 | 0.201 | 0.4981 | Yes |
| 89 | SERPING1 | NA | 2784 | 0.199 | 0.5023 | Yes |
| 90 | IGKV1-17 | NA | 2814 | 0.199 | 0.5064 | Yes |
| 91 | IGHV4-39 | NA | 2816 | 0.199 | 0.5110 | Yes |
| 92 | IGKV1D-12 | NA | 2817 | 0.199 | 0.5157 | Yes |
| 93 | IGLC1 | NA | 2899 | 0.195 | 0.5187 | Yes |
| 94 | CD27 | NA | 2927 | 0.195 | 0.5228 | Yes |
| 95 | IGHV3-43 | NA | 3029 | 0.191 | 0.5254 | Yes |
| 96 | IGHV4-28 | NA | 3075 | 0.190 | 0.5290 | Yes |
| 97 | C9 | NA | 3242 | 0.185 | 0.5303 | Yes |
| 98 | CR2 | NA | 3270 | 0.184 | 0.5341 | Yes |
| 99 | IGLV1-44 | NA | 3311 | 0.183 | 0.5376 | Yes |
| 100 | IGHV1-45 | NA | 3364 | 0.182 | 0.5409 | Yes |
| 101 | IGHV7-4-1 | NA | 3393 | 0.181 | 0.5446 | Yes |
| 102 | IGHV3-16 | NA | 3394 | 0.181 | 0.5489 | Yes |
| 103 | UNG | NA | 3485 | 0.178 | 0.5514 | Yes |
| 104 | TFRC | NA | 3506 | 0.177 | 0.5551 | Yes |
| 105 | IGHV3-53 | NA | 3723 | 0.172 | 0.5552 | Yes |
| 106 | IGHM | NA | 3785 | 0.170 | 0.5580 | Yes |
| 107 | MASP2 | NA | 3857 | 0.168 | 0.5606 | Yes |
| 108 | FCER2 | NA | 3869 | 0.168 | 0.5644 | Yes |
| 109 | TGFB1 | NA | 3942 | 0.166 | 0.5669 | Yes |
| 110 | IGHV6-1 | NA | 3991 | 0.165 | 0.5699 | Yes |
| 111 | IGHV5-10-1 | NA | 4000 | 0.164 | 0.5736 | Yes |
| 112 | IGHV4-31 | NA | 4069 | 0.163 | 0.5761 | Yes |
| 113 | IGLV6-57 | NA | 4269 | 0.158 | 0.5762 | Yes |
| 114 | IGHV3-66 | NA | 4327 | 0.156 | 0.5788 | Yes |
| 115 | C5 | NA | 4330 | 0.156 | 0.5824 | Yes |
| 116 | CD40LG | NA | 4464 | 0.153 | 0.5835 | Yes |
| 117 | NOD2 | NA | 4498 | 0.152 | 0.5865 | Yes |
| 118 | IGKV2-29 | NA | 4573 | 0.151 | 0.5886 | Yes |
| 119 | IL10 | NA | 4670 | 0.149 | 0.5903 | Yes |
| 120 | C4BPA | NA | 4672 | 0.149 | 0.5938 | Yes |
| 121 | BTK | NA | 4928 | 0.143 | 0.5924 | Yes |
| 122 | LTA | NA | 5001 | 0.142 | 0.5944 | Yes |
| 123 | CD226 | NA | 5158 | 0.139 | 0.5948 | Yes |
| 124 | CR1 | NA | 5194 | 0.139 | 0.5974 | Yes |
| 125 | HLA-G | NA | 5419 | 0.134 | 0.5964 | Yes |
| 126 | NDFIP1 | NA | 5649 | 0.130 | 0.5953 | Yes |
| 127 | CRP | NA | 5763 | 0.127 | 0.5962 | Yes |
| 128 | C1QBP | NA | 5767 | 0.127 | 0.5991 | Yes |
| 129 | C1S | NA | 5931 | 0.124 | 0.5990 | No |
| 130 | EXO1 | NA | 6298 | 0.118 | 0.5950 | No |
| 131 | CD28 | NA | 6477 | 0.115 | 0.5944 | No |
| 132 | TRBC1 | NA | 6565 | 0.114 | 0.5955 | No |
| 133 | BATF | NA | 6659 | 0.112 | 0.5964 | No |
| 134 | PRKCD | NA | 6985 | 0.107 | 0.5929 | No |
| 135 | IGHV3-64D | NA | 7005 | 0.107 | 0.5951 | No |
| 136 | IGLC7 | NA | 7419 | 0.100 | 0.5898 | No |
| 137 | TRBC2 | NA | 7436 | 0.100 | 0.5919 | No |
| 138 | CD19 | NA | 7476 | 0.099 | 0.5935 | No |
| 139 | CD74 | NA | 7865 | 0.094 | 0.5885 | No |
| 140 | IL13RA2 | NA | 7911 | 0.093 | 0.5899 | No |
| 141 | GAPT | NA | 8517 | 0.084 | 0.5807 | No |
| 142 | CCR6 | NA | 8773 | 0.081 | 0.5779 | No |
| 143 | C4A | NA | 9029 | 0.078 | 0.5750 | No |
| 144 | C17orf99 | NA | 9087 | 0.077 | 0.5758 | No |
| 145 | SWAP70 | NA | 9282 | 0.075 | 0.5740 | No |
| 146 | TRDC | NA | 10333 | 0.062 | 0.5561 | No |
| 147 | C1R | NA | 10343 | 0.062 | 0.5574 | No |
| 148 | EXOSC6 | NA | 10360 | 0.062 | 0.5585 | No |
| 149 | PTPN6 | NA | 10393 | 0.062 | 0.5594 | No |
| 150 | C6 | NA | 10692 | 0.058 | 0.5553 | No |
| 151 | RIF1 | NA | 10735 | 0.058 | 0.5559 | No |
| 152 | PTPRC | NA | 10760 | 0.058 | 0.5568 | No |
| 153 | CD70 | NA | 11750 | 0.048 | 0.5397 | No |
| 154 | C4B | NA | 12255 | 0.043 | 0.5314 | No |
| 155 | C8A | NA | 12895 | 0.038 | 0.5206 | No |
| 156 | CLCF1 | NA | 13019 | 0.037 | 0.5191 | No |
| 157 | MSH2 | NA | 13240 | 0.035 | 0.5159 | No |
| 158 | MSH6 | NA | 14774 | 0.024 | 0.4883 | No |
| 159 | C8B | NA | 15305 | 0.020 | 0.4790 | No |
| 160 | INPP5D | NA | 15626 | 0.018 | 0.4735 | No |
| 161 | HLA-DQB1 | NA | 15698 | 0.018 | 0.4726 | No |
| 162 | MLH1 | NA | 15743 | 0.017 | 0.4722 | No |
| 163 | IGHA1 | NA | 16252 | 0.015 | 0.4632 | No |
| 164 | XCL1 | NA | 18217 | 0.005 | 0.4272 | No |
| 165 | C7 | NA | 25876 | -0.007 | 0.2866 | No |
| 166 | IGHA2 | NA | 25884 | -0.007 | 0.2866 | No |
| 167 | C3 | NA | 26097 | -0.010 | 0.2830 | No |
| 168 | BCL10 | NA | 26199 | -0.011 | 0.2814 | No |
| 169 | IL27RA | NA | 26246 | -0.011 | 0.2808 | No |
| 170 | HLA-DRB1 | NA | 26700 | -0.016 | 0.2728 | No |
| 171 | KMT5B | NA | 27154 | -0.020 | 0.2649 | No |
| 172 | APCS | NA | 27223 | -0.020 | 0.2642 | No |
| 173 | SLA2 | NA | 29967 | -0.046 | 0.2148 | No |
| 174 | TBX21 | NA | 32603 | -0.073 | 0.1680 | No |
| 175 | CD55 | NA | 32929 | -0.076 | 0.1638 | No |
| 176 | TNF | NA | 33344 | -0.080 | 0.1581 | No |
| 177 | HLA-E | NA | 33665 | -0.083 | 0.1541 | No |
| 178 | TNFSF4 | NA | 33952 | -0.086 | 0.1509 | No |
| 179 | CD40 | NA | 34772 | -0.094 | 0.1380 | No |
| 180 | CLU | NA | 35202 | -0.098 | 0.1324 | No |
| 181 | ZP3 | NA | 36824 | -0.114 | 0.1053 | No |
| 182 | SLC15A4 | NA | 37395 | -0.120 | 0.0976 | No |
| 183 | MBL2 | NA | 38004 | -0.126 | 0.0894 | No |
| 184 | RNF168 | NA | 38008 | -0.126 | 0.0923 | No |
| 185 | SUPT6H | NA | 38534 | -0.132 | 0.0857 | No |
| 186 | C1RL | NA | 38743 | -0.135 | 0.0850 | No |
| 187 | FOXJ1 | NA | 38999 | -0.137 | 0.0835 | No |
| 188 | CFI | NA | 42287 | -0.177 | 0.0272 | No |
| 189 | IL2 | NA | 44270 | -0.204 | -0.0045 | No |
| 190 | IL4R | NA | 44744 | -0.211 | -0.0082 | No |
| 191 | LIG4 | NA | 45525 | -0.222 | -0.0174 | No |
| 192 | SUSD4 | NA | 45653 | -0.225 | -0.0145 | No |
| 193 | NECTIN2 | NA | 46045 | -0.231 | -0.0163 | No |
| 194 | PARP3 | NA | 46571 | -0.240 | -0.0203 | No |
| 195 | BCL3 | NA | 46892 | -0.245 | -0.0205 | No |
| 196 | PAXIP1 | NA | 47382 | -0.253 | -0.0235 | No |
| 197 | RNF8 | NA | 47709 | -0.260 | -0.0235 | No |
| 198 | NSD2 | NA | 48670 | -0.279 | -0.0346 | No |
| 199 | C8G | NA | 48988 | -0.286 | -0.0337 | No |
| 200 | EXOSC3 | NA | 49078 | -0.288 | -0.0286 | No |
| 201 | IRF7 | NA | 49193 | -0.291 | -0.0239 | No |
| 202 | TP53BP1 | NA | 49244 | -0.292 | -0.0180 | No |
| 203 | TNFSF13 | NA | 49785 | -0.304 | -0.0208 | No |
| 204 | CD81 | NA | 49796 | -0.305 | -0.0139 | No |
| 205 | HSPD1 | NA | 50170 | -0.313 | -0.0134 | No |
| 206 | HPX | NA | 50642 | -0.326 | -0.0145 | No |
| 207 | CD46 | NA | 50667 | -0.327 | -0.0073 | No |
| 208 | ERCC1 | NA | 51141 | -0.340 | -0.0080 | No |
| 209 | IGHE | NA | 51792 | -0.361 | -0.0115 | No |
| 210 | IL4 | NA | 52635 | -0.393 | -0.0179 | No |
| 211 | BCL6 | NA | 52875 | -0.404 | -0.0128 | No |
| 212 | KMT5C | NA | 53064 | -0.415 | -0.0066 | No |
| 213 | THOC1 | NA | 53802 | -0.465 | -0.0092 | No |
| 214 | TCIRG1 | NA | 53843 | -0.470 | 0.0010 | No |
| 215 | MAD2L2 | NA | 54348 | -0.545 | 0.0045 | No |