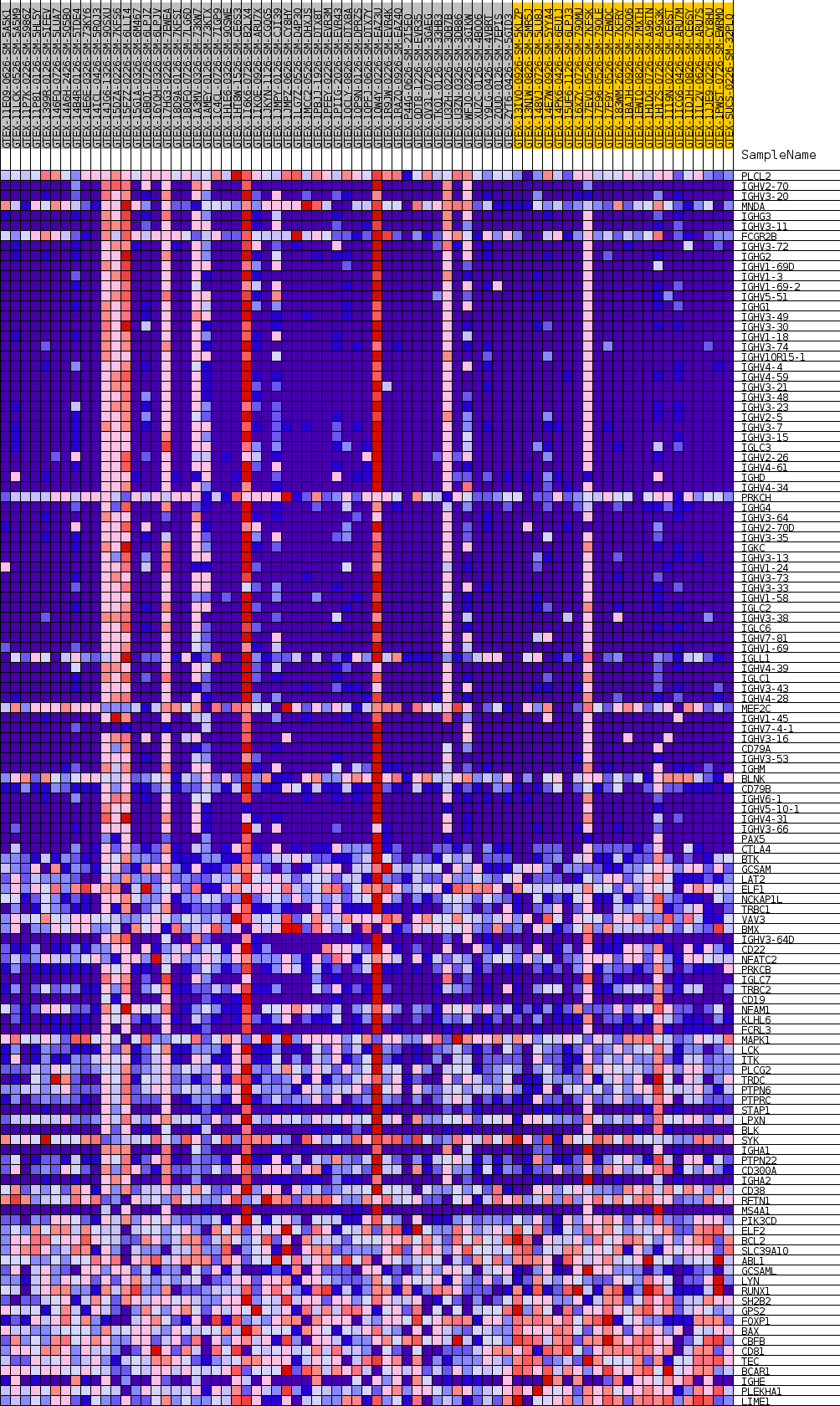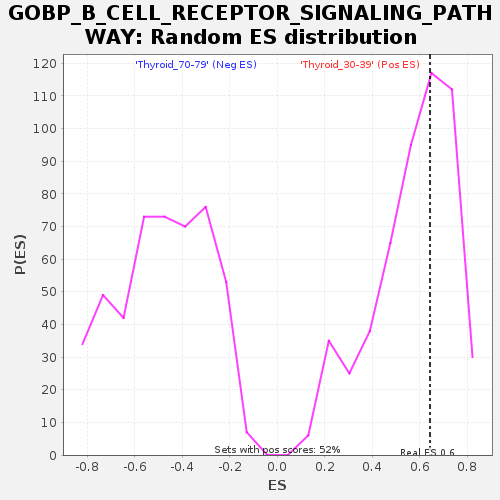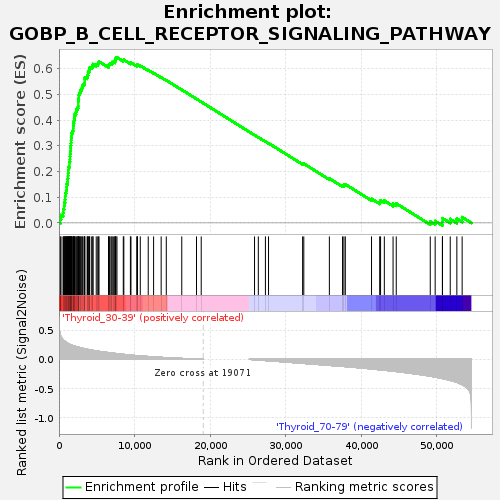
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Thyroid.Thyroid.cls #Thyroid_30-39_versus_Thyroid_70-79.Thyroid.cls #Thyroid_30-39_versus_Thyroid_70-79_repos |
| Phenotype | Thyroid.cls#Thyroid_30-39_versus_Thyroid_70-79_repos |
| Upregulated in class | Thyroid_30-39 |
| GeneSet | GOBP_B_CELL_RECEPTOR_SIGNALING_PATHWAY |
| Enrichment Score (ES) | 0.64329094 |
| Normalized Enrichment Score (NES) | 1.1273799 |
| Nominal p-value | 0.39388144 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | PLCL2 | NA | 171 | 0.418 | 0.0152 | Yes |
| 2 | IGHV2-70 | NA | 266 | 0.384 | 0.0302 | Yes |
| 3 | IGHV3-20 | NA | 541 | 0.333 | 0.0398 | Yes |
| 4 | MNDA | NA | 584 | 0.327 | 0.0533 | Yes |
| 5 | IGHG3 | NA | 685 | 0.317 | 0.0653 | Yes |
| 6 | IGHV3-11 | NA | 702 | 0.315 | 0.0788 | Yes |
| 7 | FCGR2B | NA | 799 | 0.305 | 0.0904 | Yes |
| 8 | IGHV3-72 | NA | 821 | 0.303 | 0.1033 | Yes |
| 9 | IGHG2 | NA | 846 | 0.301 | 0.1160 | Yes |
| 10 | IGHV1-69D | NA | 947 | 0.290 | 0.1269 | Yes |
| 11 | IGHV1-3 | NA | 1000 | 0.286 | 0.1384 | Yes |
| 12 | IGHV1-69-2 | NA | 1014 | 0.284 | 0.1506 | Yes |
| 13 | IGHV5-51 | NA | 1135 | 0.274 | 0.1604 | Yes |
| 14 | IGHG1 | NA | 1146 | 0.273 | 0.1722 | Yes |
| 15 | IGHV3-49 | NA | 1199 | 0.270 | 0.1830 | Yes |
| 16 | IGHV3-30 | NA | 1213 | 0.269 | 0.1945 | Yes |
| 17 | IGHV1-18 | NA | 1236 | 0.267 | 0.2058 | Yes |
| 18 | IGHV3-74 | NA | 1243 | 0.267 | 0.2174 | Yes |
| 19 | IGHV1OR15-1 | NA | 1378 | 0.258 | 0.2262 | Yes |
| 20 | IGHV4-4 | NA | 1379 | 0.258 | 0.2375 | Yes |
| 21 | IGHV4-59 | NA | 1451 | 0.254 | 0.2473 | Yes |
| 22 | IGHV3-21 | NA | 1460 | 0.253 | 0.2582 | Yes |
| 23 | IGHV3-48 | NA | 1478 | 0.252 | 0.2689 | Yes |
| 24 | IGHV3-23 | NA | 1510 | 0.251 | 0.2793 | Yes |
| 25 | IGHV2-5 | NA | 1536 | 0.249 | 0.2898 | Yes |
| 26 | IGHV3-7 | NA | 1550 | 0.248 | 0.3004 | Yes |
| 27 | IGHV3-15 | NA | 1586 | 0.246 | 0.3106 | Yes |
| 28 | IGLC3 | NA | 1602 | 0.246 | 0.3210 | Yes |
| 29 | IGHV2-26 | NA | 1612 | 0.245 | 0.3316 | Yes |
| 30 | IGHV4-61 | NA | 1690 | 0.242 | 0.3407 | Yes |
| 31 | IGHD | NA | 1697 | 0.241 | 0.3512 | Yes |
| 32 | IGHV4-34 | NA | 1818 | 0.235 | 0.3593 | Yes |
| 33 | PRKCH | NA | 1900 | 0.232 | 0.3679 | Yes |
| 34 | IGHG4 | NA | 1903 | 0.232 | 0.3780 | Yes |
| 35 | IGHV3-64 | NA | 1932 | 0.231 | 0.3876 | Yes |
| 36 | IGHV2-70D | NA | 1979 | 0.229 | 0.3968 | Yes |
| 37 | IGHV3-35 | NA | 2011 | 0.228 | 0.4062 | Yes |
| 38 | IGKC | NA | 2057 | 0.226 | 0.4152 | Yes |
| 39 | IGHV3-13 | NA | 2110 | 0.224 | 0.4241 | Yes |
| 40 | IGHV1-24 | NA | 2244 | 0.219 | 0.4312 | Yes |
| 41 | IGHV3-73 | NA | 2283 | 0.217 | 0.4400 | Yes |
| 42 | IGHV3-33 | NA | 2419 | 0.212 | 0.4468 | Yes |
| 43 | IGHV1-58 | NA | 2539 | 0.208 | 0.4538 | Yes |
| 44 | IGLC2 | NA | 2547 | 0.208 | 0.4627 | Yes |
| 45 | IGHV3-38 | NA | 2548 | 0.208 | 0.4718 | Yes |
| 46 | IGLC6 | NA | 2574 | 0.207 | 0.4804 | Yes |
| 47 | IGHV7-81 | NA | 2622 | 0.205 | 0.4885 | Yes |
| 48 | IGHV1-69 | NA | 2627 | 0.205 | 0.4974 | Yes |
| 49 | IGLL1 | NA | 2697 | 0.202 | 0.5050 | Yes |
| 50 | IGHV4-39 | NA | 2816 | 0.199 | 0.5115 | Yes |
| 51 | IGLC1 | NA | 2899 | 0.195 | 0.5186 | Yes |
| 52 | IGHV3-43 | NA | 3029 | 0.191 | 0.5245 | Yes |
| 53 | IGHV4-28 | NA | 3075 | 0.190 | 0.5320 | Yes |
| 54 | MEF2C | NA | 3185 | 0.186 | 0.5382 | Yes |
| 55 | IGHV1-45 | NA | 3364 | 0.182 | 0.5429 | Yes |
| 56 | IGHV7-4-1 | NA | 3393 | 0.181 | 0.5503 | Yes |
| 57 | IGHV3-16 | NA | 3394 | 0.181 | 0.5582 | Yes |
| 58 | CD79A | NA | 3412 | 0.180 | 0.5657 | Yes |
| 59 | IGHV3-53 | NA | 3723 | 0.172 | 0.5676 | Yes |
| 60 | IGHM | NA | 3785 | 0.170 | 0.5739 | Yes |
| 61 | BLNK | NA | 3853 | 0.168 | 0.5800 | Yes |
| 62 | CD79B | NA | 3856 | 0.168 | 0.5873 | Yes |
| 63 | IGHV6-1 | NA | 3991 | 0.165 | 0.5920 | Yes |
| 64 | IGHV5-10-1 | NA | 4000 | 0.164 | 0.5991 | Yes |
| 65 | IGHV4-31 | NA | 4069 | 0.163 | 0.6050 | Yes |
| 66 | IGHV3-66 | NA | 4327 | 0.156 | 0.6071 | Yes |
| 67 | PAX5 | NA | 4488 | 0.153 | 0.6108 | Yes |
| 68 | CTLA4 | NA | 4492 | 0.152 | 0.6174 | Yes |
| 69 | BTK | NA | 4928 | 0.143 | 0.6157 | Yes |
| 70 | GCSAM | NA | 5142 | 0.140 | 0.6179 | Yes |
| 71 | LAT2 | NA | 5195 | 0.139 | 0.6230 | Yes |
| 72 | ELF1 | NA | 5297 | 0.136 | 0.6271 | Yes |
| 73 | NCKAP1L | NA | 6555 | 0.114 | 0.6090 | Yes |
| 74 | TRBC1 | NA | 6565 | 0.114 | 0.6138 | Yes |
| 75 | VAV3 | NA | 6667 | 0.112 | 0.6169 | Yes |
| 76 | BMX | NA | 6767 | 0.110 | 0.6199 | Yes |
| 77 | IGHV3-64D | NA | 7005 | 0.107 | 0.6202 | Yes |
| 78 | CD22 | NA | 7010 | 0.107 | 0.6248 | Yes |
| 79 | NFATC2 | NA | 7189 | 0.104 | 0.6260 | Yes |
| 80 | PRKCB | NA | 7404 | 0.101 | 0.6265 | Yes |
| 81 | IGLC7 | NA | 7419 | 0.100 | 0.6306 | Yes |
| 82 | TRBC2 | NA | 7436 | 0.100 | 0.6347 | Yes |
| 83 | CD19 | NA | 7476 | 0.099 | 0.6383 | Yes |
| 84 | NFAM1 | NA | 7537 | 0.098 | 0.6415 | Yes |
| 85 | KLHL6 | NA | 7672 | 0.096 | 0.6433 | Yes |
| 86 | FCRL3 | NA | 8510 | 0.084 | 0.6316 | No |
| 87 | MAPK1 | NA | 8583 | 0.083 | 0.6339 | No |
| 88 | LCK | NA | 9451 | 0.073 | 0.6212 | No |
| 89 | ITK | NA | 9525 | 0.072 | 0.6230 | No |
| 90 | PLCG2 | NA | 10314 | 0.063 | 0.6113 | No |
| 91 | TRDC | NA | 10333 | 0.062 | 0.6137 | No |
| 92 | PTPN6 | NA | 10393 | 0.062 | 0.6153 | No |
| 93 | PTPRC | NA | 10760 | 0.058 | 0.6111 | No |
| 94 | STAP1 | NA | 11812 | 0.047 | 0.5938 | No |
| 95 | LPXN | NA | 12522 | 0.041 | 0.5826 | No |
| 96 | BLK | NA | 13523 | 0.033 | 0.5657 | No |
| 97 | SYK | NA | 14202 | 0.028 | 0.5544 | No |
| 98 | IGHA1 | NA | 16252 | 0.015 | 0.5174 | No |
| 99 | PTPN22 | NA | 18203 | 0.005 | 0.4819 | No |
| 100 | CD300A | NA | 18827 | 0.002 | 0.4706 | No |
| 101 | IGHA2 | NA | 25884 | -0.007 | 0.3413 | No |
| 102 | CD38 | NA | 26386 | -0.013 | 0.3327 | No |
| 103 | RFTN1 | NA | 27325 | -0.021 | 0.3164 | No |
| 104 | MS4A1 | NA | 27738 | -0.025 | 0.3099 | No |
| 105 | PIK3CD | NA | 32248 | -0.069 | 0.2302 | No |
| 106 | ELF2 | NA | 32406 | -0.071 | 0.2304 | No |
| 107 | BCL2 | NA | 35791 | -0.104 | 0.1728 | No |
| 108 | SLC39A10 | NA | 37534 | -0.121 | 0.1461 | No |
| 109 | ABL1 | NA | 37715 | -0.123 | 0.1482 | No |
| 110 | GCSAML | NA | 37902 | -0.125 | 0.1502 | No |
| 111 | LYN | NA | 41372 | -0.165 | 0.0938 | No |
| 112 | RUNX1 | NA | 42453 | -0.179 | 0.0818 | No |
| 113 | SH2B2 | NA | 42570 | -0.181 | 0.0875 | No |
| 114 | GPS2 | NA | 43066 | -0.187 | 0.0866 | No |
| 115 | FOXP1 | NA | 44223 | -0.203 | 0.0743 | No |
| 116 | BAX | NA | 44646 | -0.209 | 0.0757 | No |
| 117 | CBFB | NA | 49147 | -0.290 | 0.0058 | No |
| 118 | CD81 | NA | 49796 | -0.305 | 0.0072 | No |
| 119 | TEC | NA | 50755 | -0.329 | 0.0040 | No |
| 120 | BCAR1 | NA | 50764 | -0.329 | 0.0182 | No |
| 121 | IGHE | NA | 51792 | -0.361 | 0.0152 | No |
| 122 | PLEKHA1 | NA | 52667 | -0.394 | 0.0163 | No |
| 123 | LIME1 | NA | 53368 | -0.433 | 0.0225 | No |