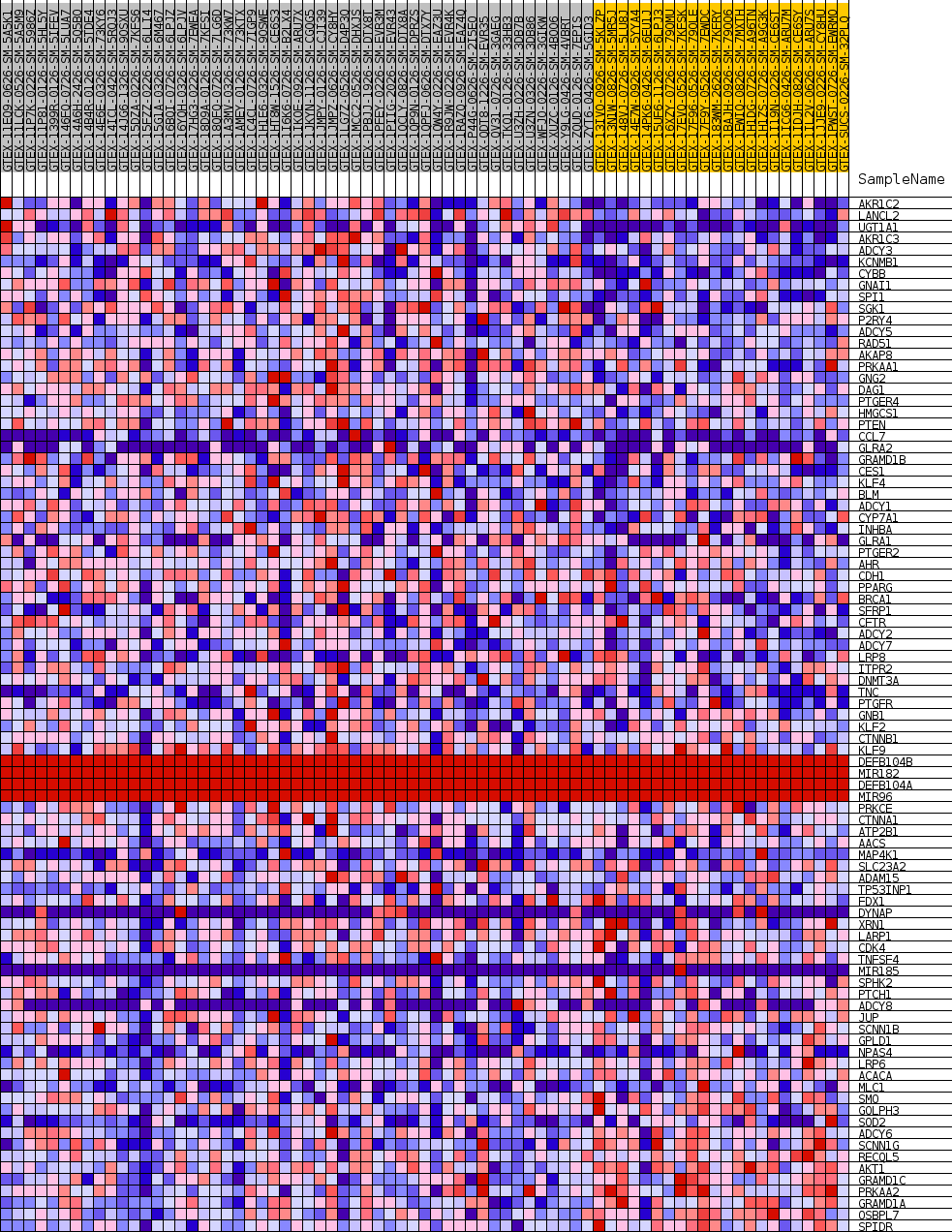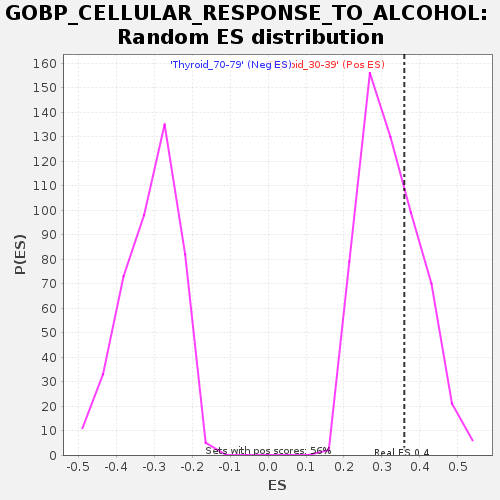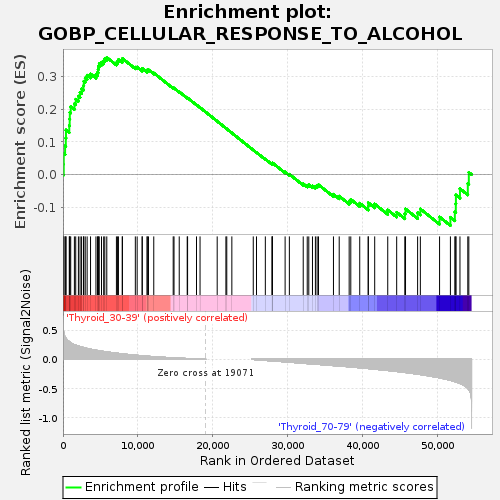
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Thyroid.Thyroid.cls #Thyroid_30-39_versus_Thyroid_70-79.Thyroid.cls #Thyroid_30-39_versus_Thyroid_70-79_repos |
| Phenotype | Thyroid.cls#Thyroid_30-39_versus_Thyroid_70-79_repos |
| Upregulated in class | Thyroid_30-39 |
| GeneSet | GOBP_CELLULAR_RESPONSE_TO_ALCOHOL |
| Enrichment Score (ES) | 0.35900536 |
| Normalized Enrichment Score (NES) | 1.1102747 |
| Nominal p-value | 0.3285968 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | AKR1C2 | NA | 95 | 0.466 | 0.0312 | Yes |
| 2 | LANCL2 | NA | 97 | 0.464 | 0.0641 | Yes |
| 3 | UGT1A1 | NA | 280 | 0.382 | 0.0879 | Yes |
| 4 | AKR1C3 | NA | 374 | 0.361 | 0.1117 | Yes |
| 5 | ADCY3 | NA | 397 | 0.356 | 0.1365 | Yes |
| 6 | KCNMB1 | NA | 839 | 0.302 | 0.1498 | Yes |
| 7 | CYBB | NA | 892 | 0.296 | 0.1698 | Yes |
| 8 | GNAI1 | NA | 909 | 0.294 | 0.1903 | Yes |
| 9 | SPI1 | NA | 1030 | 0.283 | 0.2082 | Yes |
| 10 | SGK1 | NA | 1522 | 0.250 | 0.2169 | Yes |
| 11 | P2RY4 | NA | 1699 | 0.241 | 0.2308 | Yes |
| 12 | ADCY5 | NA | 2058 | 0.226 | 0.2402 | Yes |
| 13 | RAD51 | NA | 2279 | 0.218 | 0.2516 | Yes |
| 14 | AKAP8 | NA | 2482 | 0.210 | 0.2628 | Yes |
| 15 | PRKAA1 | NA | 2737 | 0.201 | 0.2724 | Yes |
| 16 | GNG2 | NA | 2776 | 0.200 | 0.2858 | Yes |
| 17 | DAG1 | NA | 2969 | 0.193 | 0.2960 | Yes |
| 18 | PTGER4 | NA | 3225 | 0.185 | 0.3044 | Yes |
| 19 | HMGCS1 | NA | 3680 | 0.173 | 0.3083 | Yes |
| 20 | PTEN | NA | 4401 | 0.155 | 0.3061 | Yes |
| 21 | CCL7 | NA | 4608 | 0.150 | 0.3129 | Yes |
| 22 | GLRA2 | NA | 4695 | 0.148 | 0.3219 | Yes |
| 23 | GRAMD1B | NA | 4727 | 0.147 | 0.3317 | Yes |
| 24 | CES1 | NA | 4842 | 0.145 | 0.3399 | Yes |
| 25 | KLF4 | NA | 5145 | 0.139 | 0.3443 | Yes |
| 26 | BLM | NA | 5437 | 0.134 | 0.3484 | Yes |
| 27 | ADCY1 | NA | 5584 | 0.131 | 0.3550 | Yes |
| 28 | CYP7A1 | NA | 5850 | 0.126 | 0.3590 | Yes |
| 29 | INHBA | NA | 7137 | 0.105 | 0.3428 | No |
| 30 | GLRA1 | NA | 7291 | 0.102 | 0.3472 | No |
| 31 | PTGER2 | NA | 7406 | 0.101 | 0.3523 | No |
| 32 | AHR | NA | 7913 | 0.093 | 0.3496 | No |
| 33 | CDH1 | NA | 7944 | 0.093 | 0.3556 | No |
| 34 | PPARG | NA | 9664 | 0.070 | 0.3290 | No |
| 35 | BRCA1 | NA | 9908 | 0.067 | 0.3293 | No |
| 36 | SFRP1 | NA | 10580 | 0.060 | 0.3212 | No |
| 37 | CFTR | NA | 10584 | 0.060 | 0.3254 | No |
| 38 | ADCY2 | NA | 11237 | 0.053 | 0.3172 | No |
| 39 | ADCY7 | NA | 11256 | 0.053 | 0.3206 | No |
| 40 | LRP8 | NA | 11421 | 0.051 | 0.3212 | No |
| 41 | ITPR2 | NA | 12127 | 0.044 | 0.3113 | No |
| 42 | DNMT3A | NA | 14723 | 0.024 | 0.2654 | No |
| 43 | TNC | NA | 14833 | 0.023 | 0.2651 | No |
| 44 | PTGFR | NA | 15530 | 0.019 | 0.2536 | No |
| 45 | GNB1 | NA | 16596 | 0.013 | 0.2350 | No |
| 46 | KLF2 | NA | 16647 | 0.012 | 0.2350 | No |
| 47 | CTNNB1 | NA | 17837 | 0.007 | 0.2136 | No |
| 48 | KLF9 | NA | 18311 | 0.005 | 0.2053 | No |
| 49 | DEFB104B | NA | 20602 | 0.000 | 0.1633 | No |
| 50 | MIR182 | NA | 21766 | 0.000 | 0.1420 | No |
| 51 | DEFB104A | NA | 21882 | 0.000 | 0.1398 | No |
| 52 | MIR96 | NA | 22560 | 0.000 | 0.1274 | No |
| 53 | PRKCE | NA | 25421 | -0.003 | 0.0751 | No |
| 54 | CTNNA1 | NA | 25853 | -0.007 | 0.0677 | No |
| 55 | ATP2B1 | NA | 27034 | -0.018 | 0.0474 | No |
| 56 | AACS | NA | 27929 | -0.027 | 0.0329 | No |
| 57 | MAP4K1 | NA | 27955 | -0.027 | 0.0343 | No |
| 58 | SLC23A2 | NA | 27989 | -0.027 | 0.0357 | No |
| 59 | ADAM15 | NA | 29677 | -0.043 | 0.0078 | No |
| 60 | TP53INP1 | NA | 30245 | -0.049 | 0.0009 | No |
| 61 | FDX1 | NA | 32097 | -0.068 | -0.0283 | No |
| 62 | DYNAP | NA | 32633 | -0.073 | -0.0329 | No |
| 63 | XRN1 | NA | 32840 | -0.075 | -0.0314 | No |
| 64 | LARP1 | NA | 33333 | -0.080 | -0.0347 | No |
| 65 | CDK4 | NA | 33724 | -0.083 | -0.0360 | No |
| 66 | TNFSF4 | NA | 33952 | -0.086 | -0.0341 | No |
| 67 | MIR185 | NA | 34136 | -0.087 | -0.0313 | No |
| 68 | SPHK2 | NA | 36137 | -0.107 | -0.0604 | No |
| 69 | PTCH1 | NA | 36905 | -0.115 | -0.0663 | No |
| 70 | ADCY8 | NA | 38235 | -0.129 | -0.0816 | No |
| 71 | JUP | NA | 38453 | -0.131 | -0.0762 | No |
| 72 | SCNN1B | NA | 39647 | -0.144 | -0.0879 | No |
| 73 | GPLD1 | NA | 40783 | -0.158 | -0.0975 | No |
| 74 | NPAS4 | NA | 40788 | -0.158 | -0.0864 | No |
| 75 | LRP6 | NA | 41647 | -0.169 | -0.0902 | No |
| 76 | ACACA | NA | 43390 | -0.191 | -0.1087 | No |
| 77 | MLC1 | NA | 44584 | -0.208 | -0.1158 | No |
| 78 | SMO | NA | 45671 | -0.225 | -0.1198 | No |
| 79 | GOLPH3 | NA | 45766 | -0.226 | -0.1055 | No |
| 80 | SOD2 | NA | 47381 | -0.253 | -0.1171 | No |
| 81 | ADCY6 | NA | 47747 | -0.260 | -0.1054 | No |
| 82 | SCNN1G | NA | 50315 | -0.317 | -0.1300 | No |
| 83 | RECQL5 | NA | 51763 | -0.360 | -0.1310 | No |
| 84 | AKT1 | NA | 52354 | -0.381 | -0.1148 | No |
| 85 | GRAMD1C | NA | 52483 | -0.386 | -0.0898 | No |
| 86 | PRKAA2 | NA | 52498 | -0.387 | -0.0627 | No |
| 87 | GRAMD1A | NA | 53040 | -0.414 | -0.0433 | No |
| 88 | OSBPL7 | NA | 54087 | -0.496 | -0.0273 | No |
| 89 | SPIDR | NA | 54223 | -0.516 | 0.0068 | No |