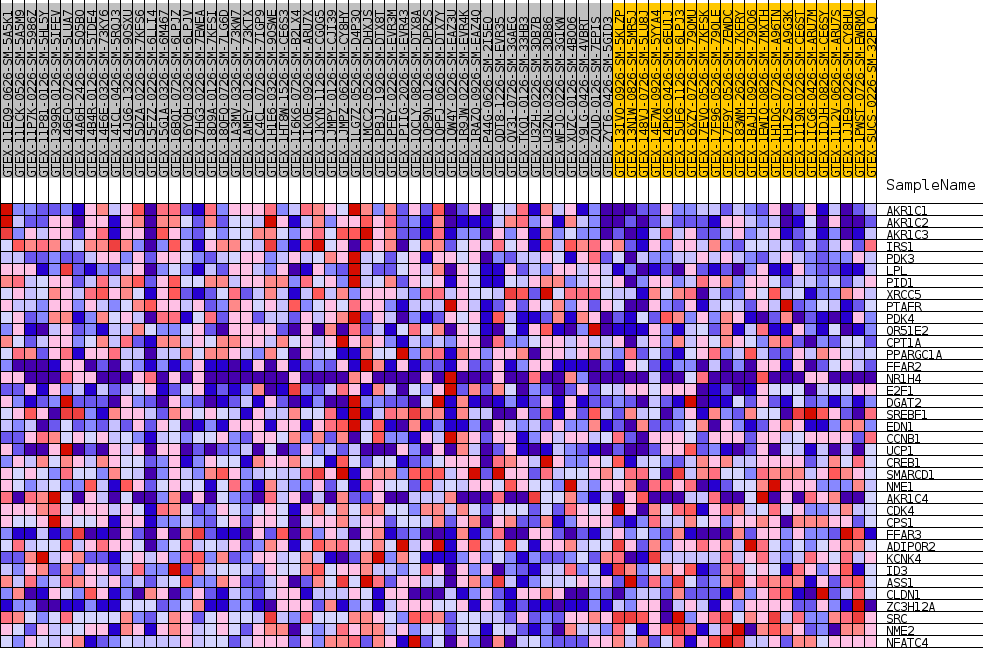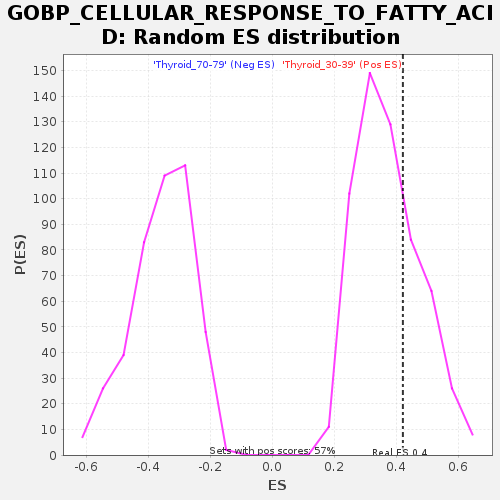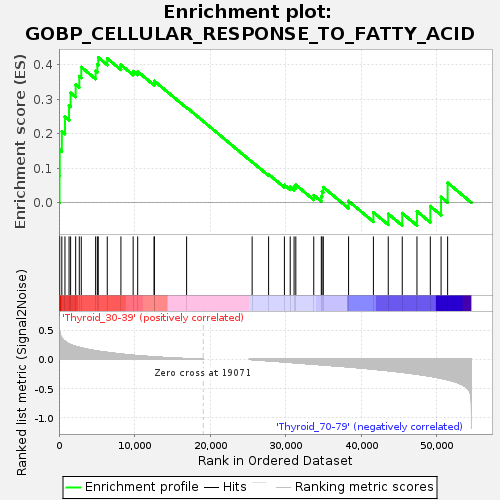
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Thyroid.Thyroid.cls #Thyroid_30-39_versus_Thyroid_70-79.Thyroid.cls #Thyroid_30-39_versus_Thyroid_70-79_repos |
| Phenotype | Thyroid.cls#Thyroid_30-39_versus_Thyroid_70-79_repos |
| Upregulated in class | Thyroid_30-39 |
| GeneSet | GOBP_CELLULAR_RESPONSE_TO_FATTY_ACID |
| Enrichment Score (ES) | 0.42138308 |
| Normalized Enrichment Score (NES) | 1.1294944 |
| Nominal p-value | 0.30017453 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | AKR1C1 | NA | 67 | 0.486 | 0.0780 | Yes |
| 2 | AKR1C2 | NA | 95 | 0.466 | 0.1533 | Yes |
| 3 | AKR1C3 | NA | 374 | 0.361 | 0.2069 | Yes |
| 4 | IRS1 | NA | 787 | 0.306 | 0.2493 | Yes |
| 5 | PDK3 | NA | 1331 | 0.261 | 0.2818 | Yes |
| 6 | LPL | NA | 1537 | 0.249 | 0.3187 | Yes |
| 7 | PID1 | NA | 2206 | 0.221 | 0.3424 | Yes |
| 8 | XRCC5 | NA | 2685 | 0.203 | 0.3667 | Yes |
| 9 | PTAFR | NA | 2949 | 0.194 | 0.3934 | Yes |
| 10 | PDK4 | NA | 4849 | 0.145 | 0.3822 | Yes |
| 11 | OR51E2 | NA | 5079 | 0.141 | 0.4010 | Yes |
| 12 | CPT1A | NA | 5196 | 0.139 | 0.4214 | Yes |
| 13 | PPARGC1A | NA | 6388 | 0.117 | 0.4185 | No |
| 14 | FFAR2 | NA | 8196 | 0.089 | 0.3999 | No |
| 15 | NR1H4 | NA | 9816 | 0.068 | 0.3814 | No |
| 16 | E2F1 | NA | 10416 | 0.061 | 0.3804 | No |
| 17 | DGAT2 | NA | 12585 | 0.040 | 0.3472 | No |
| 18 | SREBF1 | NA | 12628 | 0.040 | 0.3528 | No |
| 19 | EDN1 | NA | 16892 | 0.011 | 0.2765 | No |
| 20 | CCNB1 | NA | 25567 | -0.004 | 0.1182 | No |
| 21 | UCP1 | NA | 27746 | -0.025 | 0.0824 | No |
| 22 | CREB1 | NA | 29839 | -0.045 | 0.0514 | No |
| 23 | SMARCD1 | NA | 30608 | -0.053 | 0.0459 | No |
| 24 | NME1 | NA | 31120 | -0.058 | 0.0459 | No |
| 25 | AKR1C4 | NA | 31350 | -0.060 | 0.0516 | No |
| 26 | CDK4 | NA | 33724 | -0.083 | 0.0216 | No |
| 27 | CPS1 | NA | 34723 | -0.094 | 0.0186 | No |
| 28 | FFAR3 | NA | 34823 | -0.095 | 0.0322 | No |
| 29 | ADIPOR2 | NA | 34990 | -0.096 | 0.0448 | No |
| 30 | KCNK4 | NA | 38333 | -0.130 | 0.0047 | No |
| 31 | ID3 | NA | 41621 | -0.168 | -0.0281 | No |
| 32 | ASS1 | NA | 43586 | -0.194 | -0.0326 | No |
| 33 | CLDN1 | NA | 45440 | -0.221 | -0.0306 | No |
| 34 | ZC3H12A | NA | 47386 | -0.254 | -0.0250 | No |
| 35 | SRC | NA | 49154 | -0.290 | -0.0101 | No |
| 36 | NME2 | NA | 50579 | -0.324 | 0.0166 | No |
| 37 | NFATC4 | NA | 51446 | -0.350 | 0.0576 | No |