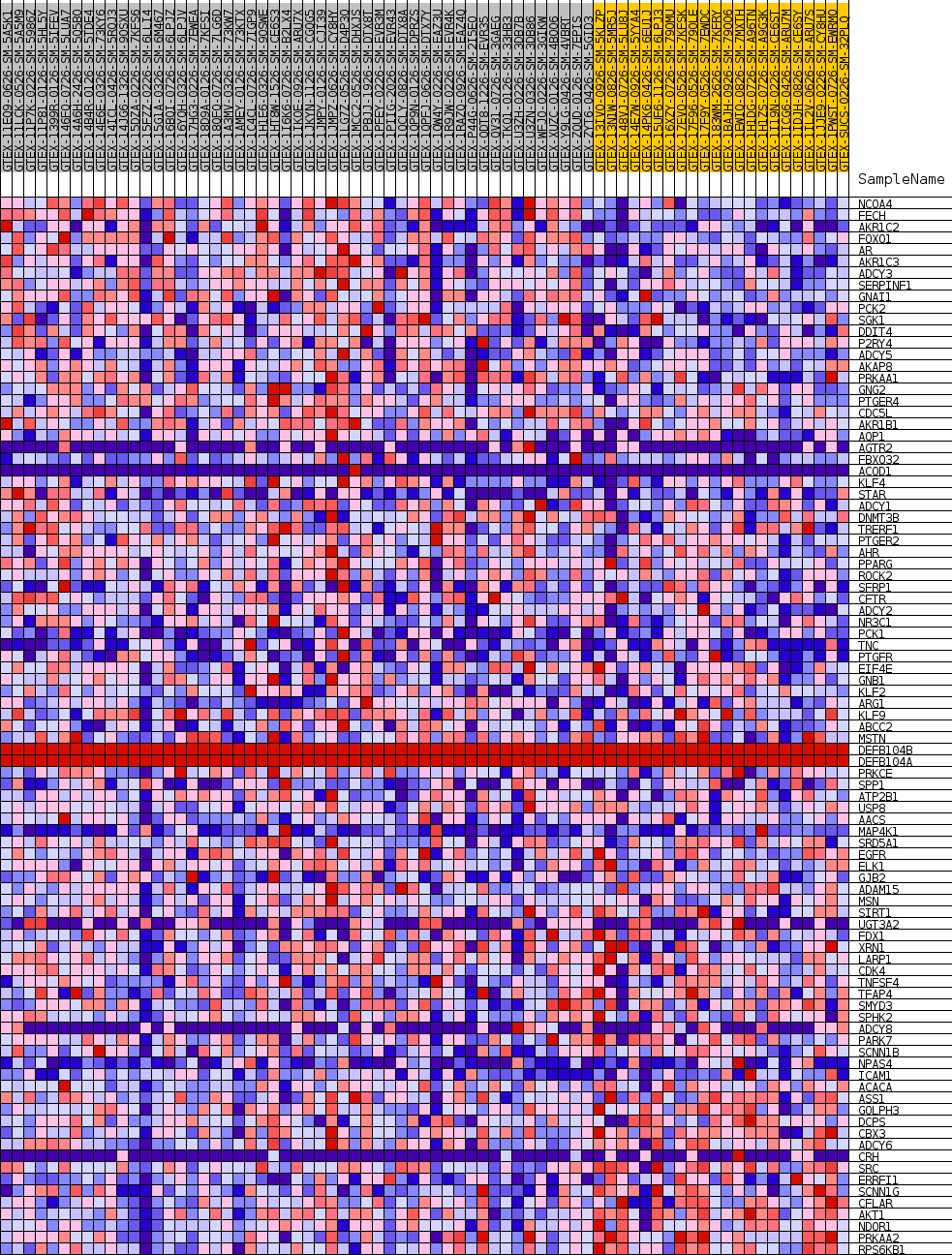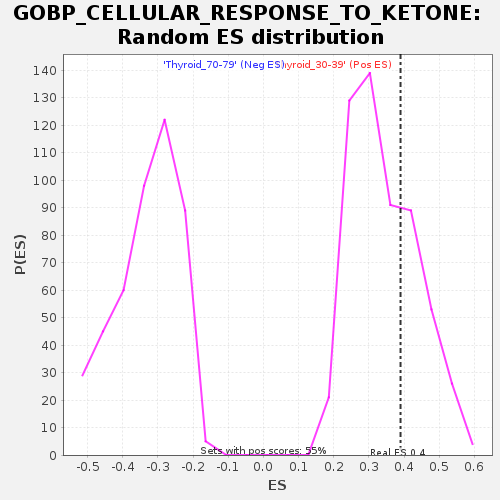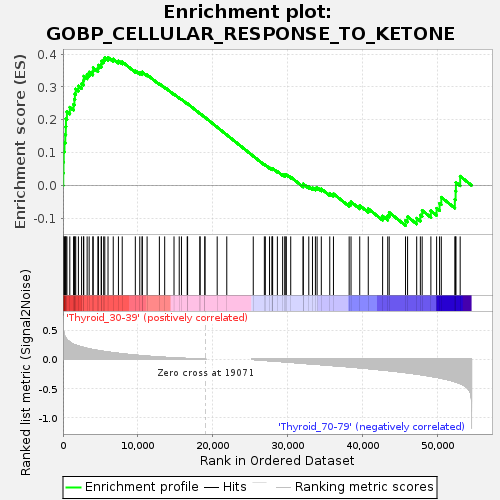
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Thyroid.Thyroid.cls #Thyroid_30-39_versus_Thyroid_70-79.Thyroid.cls #Thyroid_30-39_versus_Thyroid_70-79_repos |
| Phenotype | Thyroid.cls#Thyroid_30-39_versus_Thyroid_70-79_repos |
| Upregulated in class | Thyroid_30-39 |
| GeneSet | GOBP_CELLULAR_RESPONSE_TO_KETONE |
| Enrichment Score (ES) | 0.39004284 |
| Normalized Enrichment Score (NES) | 1.1378458 |
| Nominal p-value | 0.3115942 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | NCOA4 | NA | 32 | 0.536 | 0.0371 | Yes |
| 2 | FECH | NA | 62 | 0.490 | 0.0711 | Yes |
| 3 | AKR1C2 | NA | 95 | 0.466 | 0.1032 | Yes |
| 4 | FOXO1 | NA | 196 | 0.407 | 0.1300 | Yes |
| 5 | AR | NA | 302 | 0.376 | 0.1546 | Yes |
| 6 | AKR1C3 | NA | 374 | 0.361 | 0.1786 | Yes |
| 7 | ADCY3 | NA | 397 | 0.356 | 0.2033 | Yes |
| 8 | SERPINF1 | NA | 523 | 0.335 | 0.2246 | Yes |
| 9 | GNAI1 | NA | 909 | 0.294 | 0.2382 | Yes |
| 10 | PCK2 | NA | 1411 | 0.256 | 0.2470 | Yes |
| 11 | SGK1 | NA | 1522 | 0.250 | 0.2626 | Yes |
| 12 | DDIT4 | NA | 1589 | 0.246 | 0.2787 | Yes |
| 13 | P2RY4 | NA | 1699 | 0.241 | 0.2937 | Yes |
| 14 | ADCY5 | NA | 2058 | 0.226 | 0.3030 | Yes |
| 15 | AKAP8 | NA | 2482 | 0.210 | 0.3100 | Yes |
| 16 | PRKAA1 | NA | 2737 | 0.201 | 0.3195 | Yes |
| 17 | GNG2 | NA | 2776 | 0.200 | 0.3329 | Yes |
| 18 | PTGER4 | NA | 3225 | 0.185 | 0.3377 | Yes |
| 19 | CDC5L | NA | 3494 | 0.178 | 0.3453 | Yes |
| 20 | AKR1B1 | NA | 3984 | 0.165 | 0.3479 | Yes |
| 21 | AQP1 | NA | 4034 | 0.163 | 0.3585 | Yes |
| 22 | AGTR2 | NA | 4663 | 0.149 | 0.3575 | Yes |
| 23 | FBXO32 | NA | 4739 | 0.147 | 0.3664 | Yes |
| 24 | ACOD1 | NA | 5098 | 0.140 | 0.3697 | Yes |
| 25 | KLF4 | NA | 5145 | 0.139 | 0.3787 | Yes |
| 26 | STAR | NA | 5435 | 0.134 | 0.3828 | Yes |
| 27 | ADCY1 | NA | 5584 | 0.131 | 0.3893 | Yes |
| 28 | DNMT3B | NA | 6015 | 0.123 | 0.3900 | Yes |
| 29 | TRERF1 | NA | 6713 | 0.111 | 0.3851 | No |
| 30 | PTGER2 | NA | 7406 | 0.101 | 0.3795 | No |
| 31 | AHR | NA | 7913 | 0.093 | 0.3767 | No |
| 32 | PPARG | NA | 9664 | 0.070 | 0.3496 | No |
| 33 | ROCK2 | NA | 10257 | 0.063 | 0.3431 | No |
| 34 | SFRP1 | NA | 10580 | 0.060 | 0.3414 | No |
| 35 | CFTR | NA | 10584 | 0.060 | 0.3456 | No |
| 36 | ADCY2 | NA | 11237 | 0.053 | 0.3373 | No |
| 37 | NR3C1 | NA | 12877 | 0.038 | 0.3099 | No |
| 38 | PCK1 | NA | 13586 | 0.032 | 0.2992 | No |
| 39 | TNC | NA | 14833 | 0.023 | 0.2779 | No |
| 40 | PTGFR | NA | 15530 | 0.019 | 0.2665 | No |
| 41 | EIF4E | NA | 15828 | 0.017 | 0.2622 | No |
| 42 | GNB1 | NA | 16596 | 0.013 | 0.2490 | No |
| 43 | KLF2 | NA | 16647 | 0.012 | 0.2490 | No |
| 44 | ARG1 | NA | 18293 | 0.005 | 0.2192 | No |
| 45 | KLF9 | NA | 18311 | 0.005 | 0.2192 | No |
| 46 | ABCC2 | NA | 18953 | 0.001 | 0.2075 | No |
| 47 | MSTN | NA | 18964 | 0.001 | 0.2075 | No |
| 48 | DEFB104B | NA | 20602 | 0.000 | 0.1774 | No |
| 49 | DEFB104A | NA | 21882 | 0.000 | 0.1540 | No |
| 50 | PRKCE | NA | 25421 | -0.003 | 0.0892 | No |
| 51 | SPP1 | NA | 26904 | -0.017 | 0.0632 | No |
| 52 | ATP2B1 | NA | 27034 | -0.018 | 0.0622 | No |
| 53 | USP8 | NA | 27596 | -0.024 | 0.0536 | No |
| 54 | AACS | NA | 27929 | -0.027 | 0.0493 | No |
| 55 | MAP4K1 | NA | 27955 | -0.027 | 0.0508 | No |
| 56 | SRD5A1 | NA | 28010 | -0.028 | 0.0517 | No |
| 57 | EGFR | NA | 28643 | -0.033 | 0.0425 | No |
| 58 | ELK1 | NA | 29347 | -0.040 | 0.0324 | No |
| 59 | GJB2 | NA | 29645 | -0.043 | 0.0300 | No |
| 60 | ADAM15 | NA | 29677 | -0.043 | 0.0325 | No |
| 61 | MSN | NA | 29840 | -0.045 | 0.0327 | No |
| 62 | SIRT1 | NA | 30427 | -0.051 | 0.0255 | No |
| 63 | UGT3A2 | NA | 32084 | -0.068 | -0.0001 | No |
| 64 | FDX1 | NA | 32097 | -0.068 | 0.0044 | No |
| 65 | XRN1 | NA | 32840 | -0.075 | -0.0039 | No |
| 66 | LARP1 | NA | 33333 | -0.080 | -0.0073 | No |
| 67 | CDK4 | NA | 33724 | -0.083 | -0.0086 | No |
| 68 | TNFSF4 | NA | 33952 | -0.086 | -0.0067 | No |
| 69 | TFAP4 | NA | 34514 | -0.091 | -0.0106 | No |
| 70 | SMYD3 | NA | 35643 | -0.102 | -0.0241 | No |
| 71 | SPHK2 | NA | 36137 | -0.107 | -0.0256 | No |
| 72 | ADCY8 | NA | 38235 | -0.129 | -0.0550 | No |
| 73 | PARK7 | NA | 38485 | -0.132 | -0.0503 | No |
| 74 | SCNN1B | NA | 39647 | -0.144 | -0.0614 | No |
| 75 | NPAS4 | NA | 40788 | -0.158 | -0.0712 | No |
| 76 | ICAM1 | NA | 42705 | -0.182 | -0.0936 | No |
| 77 | ACACA | NA | 43390 | -0.191 | -0.0927 | No |
| 78 | ASS1 | NA | 43586 | -0.194 | -0.0826 | No |
| 79 | GOLPH3 | NA | 45766 | -0.226 | -0.1067 | No |
| 80 | DCPS | NA | 46041 | -0.231 | -0.0955 | No |
| 81 | CBX3 | NA | 47258 | -0.252 | -0.1001 | No |
| 82 | ADCY6 | NA | 47747 | -0.260 | -0.0907 | No |
| 83 | CRH | NA | 47989 | -0.265 | -0.0765 | No |
| 84 | SRC | NA | 49154 | -0.290 | -0.0774 | No |
| 85 | ERRFI1 | NA | 49920 | -0.308 | -0.0698 | No |
| 86 | SCNN1G | NA | 50315 | -0.317 | -0.0547 | No |
| 87 | CFLAR | NA | 50546 | -0.323 | -0.0362 | No |
| 88 | AKT1 | NA | 52354 | -0.381 | -0.0425 | No |
| 89 | NDOR1 | NA | 52458 | -0.385 | -0.0173 | No |
| 90 | PRKAA2 | NA | 52498 | -0.387 | 0.0092 | No |
| 91 | RPS6KB1 | NA | 53068 | -0.415 | 0.0279 | No |