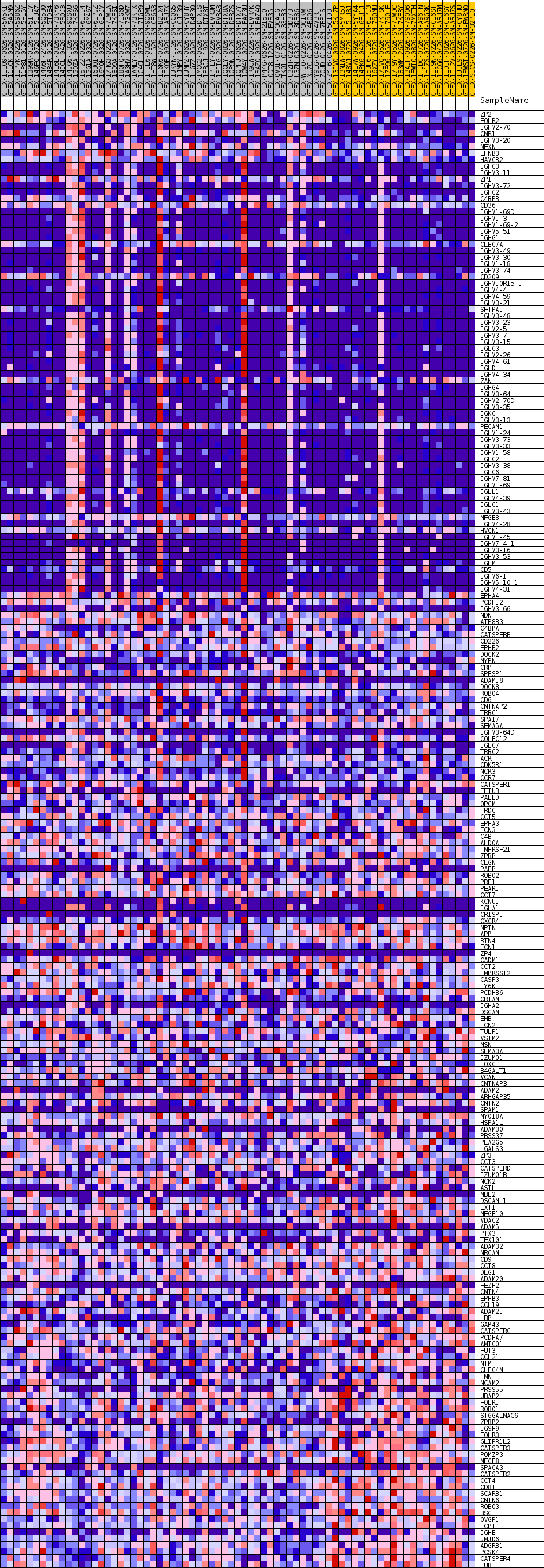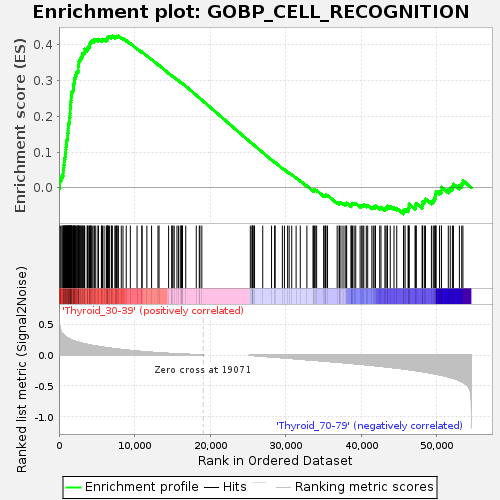
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Thyroid.Thyroid.cls #Thyroid_30-39_versus_Thyroid_70-79.Thyroid.cls #Thyroid_30-39_versus_Thyroid_70-79_repos |
| Phenotype | Thyroid.cls#Thyroid_30-39_versus_Thyroid_70-79_repos |
| Upregulated in class | Thyroid_30-39 |
| GeneSet | GOBP_CELL_RECOGNITION |
| Enrichment Score (ES) | 0.42487577 |
| Normalized Enrichment Score (NES) | 1.1228975 |
| Nominal p-value | 0.3843594 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | ZP2 | NA | 118 | 0.446 | 0.0093 | Yes |
| 2 | FOLR2 | NA | 119 | 0.446 | 0.0208 | Yes |
| 3 | IGHV2-70 | NA | 266 | 0.384 | 0.0281 | Yes |
| 4 | CNR1 | NA | 411 | 0.354 | 0.0345 | Yes |
| 5 | IGHV3-20 | NA | 541 | 0.333 | 0.0408 | Yes |
| 6 | NEXN | NA | 553 | 0.331 | 0.0491 | Yes |
| 7 | EFNB3 | NA | 615 | 0.323 | 0.0563 | Yes |
| 8 | HAVCR2 | NA | 641 | 0.321 | 0.0641 | Yes |
| 9 | IGHG3 | NA | 685 | 0.317 | 0.0715 | Yes |
| 10 | IGHV3-11 | NA | 702 | 0.315 | 0.0794 | Yes |
| 11 | ZP1 | NA | 789 | 0.306 | 0.0857 | Yes |
| 12 | IGHV3-72 | NA | 821 | 0.303 | 0.0929 | Yes |
| 13 | IGHG2 | NA | 846 | 0.301 | 0.1003 | Yes |
| 14 | C4BPB | NA | 906 | 0.294 | 0.1068 | Yes |
| 15 | CD36 | NA | 926 | 0.292 | 0.1139 | Yes |
| 16 | IGHV1-69D | NA | 947 | 0.290 | 0.1211 | Yes |
| 17 | IGHV1-3 | NA | 1000 | 0.286 | 0.1275 | Yes |
| 18 | IGHV1-69-2 | NA | 1014 | 0.284 | 0.1346 | Yes |
| 19 | IGHV5-51 | NA | 1135 | 0.274 | 0.1394 | Yes |
| 20 | IGHG1 | NA | 1146 | 0.273 | 0.1463 | Yes |
| 21 | CLEC7A | NA | 1154 | 0.273 | 0.1532 | Yes |
| 22 | IGHV3-49 | NA | 1199 | 0.270 | 0.1594 | Yes |
| 23 | IGHV3-30 | NA | 1213 | 0.269 | 0.1661 | Yes |
| 24 | IGHV1-18 | NA | 1236 | 0.267 | 0.1726 | Yes |
| 25 | IGHV3-74 | NA | 1243 | 0.267 | 0.1793 | Yes |
| 26 | CD209 | NA | 1345 | 0.260 | 0.1842 | Yes |
| 27 | IGHV1OR15-1 | NA | 1378 | 0.258 | 0.1903 | Yes |
| 28 | IGHV4-4 | NA | 1379 | 0.258 | 0.1969 | Yes |
| 29 | IGHV4-59 | NA | 1451 | 0.254 | 0.2022 | Yes |
| 30 | IGHV3-21 | NA | 1460 | 0.253 | 0.2085 | Yes |
| 31 | SFTPA1 | NA | 1475 | 0.252 | 0.2148 | Yes |
| 32 | IGHV3-48 | NA | 1478 | 0.252 | 0.2213 | Yes |
| 33 | IGHV3-23 | NA | 1510 | 0.251 | 0.2272 | Yes |
| 34 | IGHV2-5 | NA | 1536 | 0.249 | 0.2331 | Yes |
| 35 | IGHV3-7 | NA | 1550 | 0.248 | 0.2393 | Yes |
| 36 | IGHV3-15 | NA | 1586 | 0.246 | 0.2450 | Yes |
| 37 | IGLC3 | NA | 1602 | 0.246 | 0.2511 | Yes |
| 38 | IGHV2-26 | NA | 1612 | 0.245 | 0.2572 | Yes |
| 39 | IGHV4-61 | NA | 1690 | 0.242 | 0.2621 | Yes |
| 40 | IGHD | NA | 1697 | 0.241 | 0.2682 | Yes |
| 41 | IGHV4-34 | NA | 1818 | 0.235 | 0.2720 | Yes |
| 42 | ZAN | NA | 1892 | 0.232 | 0.2767 | Yes |
| 43 | IGHG4 | NA | 1903 | 0.232 | 0.2825 | Yes |
| 44 | IGHV3-64 | NA | 1932 | 0.231 | 0.2879 | Yes |
| 45 | IGHV2-70D | NA | 1979 | 0.229 | 0.2930 | Yes |
| 46 | IGHV3-35 | NA | 2011 | 0.228 | 0.2983 | Yes |
| 47 | IGKC | NA | 2057 | 0.226 | 0.3033 | Yes |
| 48 | IGHV3-13 | NA | 2110 | 0.224 | 0.3081 | Yes |
| 49 | PECAM1 | NA | 2197 | 0.221 | 0.3122 | Yes |
| 50 | IGHV1-24 | NA | 2244 | 0.219 | 0.3170 | Yes |
| 51 | IGHV3-73 | NA | 2283 | 0.217 | 0.3220 | Yes |
| 52 | IGHV3-33 | NA | 2419 | 0.212 | 0.3249 | Yes |
| 53 | IGHV1-58 | NA | 2539 | 0.208 | 0.3281 | Yes |
| 54 | IGLC2 | NA | 2547 | 0.208 | 0.3334 | Yes |
| 55 | IGHV3-38 | NA | 2548 | 0.208 | 0.3387 | Yes |
| 56 | IGLC6 | NA | 2574 | 0.207 | 0.3436 | Yes |
| 57 | IGHV7-81 | NA | 2622 | 0.205 | 0.3480 | Yes |
| 58 | IGHV1-69 | NA | 2627 | 0.205 | 0.3532 | Yes |
| 59 | IGLL1 | NA | 2697 | 0.202 | 0.3572 | Yes |
| 60 | IGHV4-39 | NA | 2816 | 0.199 | 0.3601 | Yes |
| 61 | IGLC1 | NA | 2899 | 0.195 | 0.3637 | Yes |
| 62 | IGHV3-43 | NA | 3029 | 0.191 | 0.3662 | Yes |
| 63 | MFGE8 | NA | 3057 | 0.190 | 0.3706 | Yes |
| 64 | IGHV4-28 | NA | 3075 | 0.190 | 0.3752 | Yes |
| 65 | HVCN1 | NA | 3227 | 0.185 | 0.3772 | Yes |
| 66 | IGHV1-45 | NA | 3364 | 0.182 | 0.3794 | Yes |
| 67 | IGHV7-4-1 | NA | 3393 | 0.181 | 0.3836 | Yes |
| 68 | IGHV3-16 | NA | 3394 | 0.181 | 0.3882 | Yes |
| 69 | IGHV3-53 | NA | 3723 | 0.172 | 0.3866 | Yes |
| 70 | IGHM | NA | 3785 | 0.170 | 0.3899 | Yes |
| 71 | CD5 | NA | 3825 | 0.169 | 0.3935 | Yes |
| 72 | IGHV6-1 | NA | 3991 | 0.165 | 0.3947 | Yes |
| 73 | IGHV5-10-1 | NA | 4000 | 0.164 | 0.3988 | Yes |
| 74 | IGHV4-31 | NA | 4069 | 0.163 | 0.4018 | Yes |
| 75 | EPHA4 | NA | 4128 | 0.161 | 0.4049 | Yes |
| 76 | PCDH12 | NA | 4169 | 0.160 | 0.4083 | Yes |
| 77 | IGHV3-66 | NA | 4327 | 0.156 | 0.4094 | Yes |
| 78 | NDN | NA | 4396 | 0.155 | 0.4122 | Yes |
| 79 | ATP8B3 | NA | 4636 | 0.149 | 0.4116 | Yes |
| 80 | C4BPA | NA | 4672 | 0.149 | 0.4148 | Yes |
| 81 | CATSPERB | NA | 4863 | 0.145 | 0.4150 | Yes |
| 82 | CD226 | NA | 5158 | 0.139 | 0.4132 | Yes |
| 83 | EPHB2 | NA | 5222 | 0.138 | 0.4156 | Yes |
| 84 | DOCK2 | NA | 5612 | 0.130 | 0.4118 | Yes |
| 85 | MYPN | NA | 5641 | 0.130 | 0.4147 | Yes |
| 86 | CRP | NA | 5763 | 0.127 | 0.4157 | Yes |
| 87 | SPESP1 | NA | 5965 | 0.124 | 0.4152 | Yes |
| 88 | ADAM18 | NA | 6258 | 0.119 | 0.4129 | Yes |
| 89 | DOCK8 | NA | 6305 | 0.118 | 0.4151 | Yes |
| 90 | ROBO4 | NA | 6438 | 0.116 | 0.4157 | Yes |
| 91 | CD6 | NA | 6439 | 0.116 | 0.4186 | Yes |
| 92 | CNTNAP2 | NA | 6454 | 0.116 | 0.4214 | Yes |
| 93 | TRBC1 | NA | 6565 | 0.114 | 0.4223 | Yes |
| 94 | SPA17 | NA | 6696 | 0.111 | 0.4228 | Yes |
| 95 | SEMA5A | NA | 6935 | 0.108 | 0.4212 | Yes |
| 96 | IGHV3-64D | NA | 7005 | 0.107 | 0.4226 | Yes |
| 97 | COLEC12 | NA | 7051 | 0.106 | 0.4245 | Yes |
| 98 | IGLC7 | NA | 7419 | 0.100 | 0.4204 | Yes |
| 99 | TRBC2 | NA | 7436 | 0.100 | 0.4227 | Yes |
| 100 | ACR | NA | 7598 | 0.098 | 0.4222 | Yes |
| 101 | CDK5R1 | NA | 7627 | 0.097 | 0.4242 | Yes |
| 102 | NCR3 | NA | 7788 | 0.095 | 0.4237 | Yes |
| 103 | CCR7 | NA | 7857 | 0.094 | 0.4249 | Yes |
| 104 | CATSPER1 | NA | 8243 | 0.088 | 0.4201 | No |
| 105 | FETUB | NA | 8471 | 0.085 | 0.4181 | No |
| 106 | PALLD | NA | 8901 | 0.079 | 0.4122 | No |
| 107 | OPCML | NA | 9443 | 0.073 | 0.4042 | No |
| 108 | TRDC | NA | 10333 | 0.062 | 0.3894 | No |
| 109 | CCT5 | NA | 10971 | 0.056 | 0.3791 | No |
| 110 | EPHA3 | NA | 10987 | 0.055 | 0.3803 | No |
| 111 | FCN3 | NA | 11632 | 0.049 | 0.3697 | No |
| 112 | C4B | NA | 12255 | 0.043 | 0.3594 | No |
| 113 | ALDOA | NA | 13104 | 0.036 | 0.3447 | No |
| 114 | TNFRSF21 | NA | 13245 | 0.035 | 0.3430 | No |
| 115 | ZPBP | NA | 14510 | 0.025 | 0.3204 | No |
| 116 | CLGN | NA | 14935 | 0.023 | 0.3132 | No |
| 117 | PAEP | NA | 14953 | 0.023 | 0.3135 | No |
| 118 | ROBO2 | NA | 15084 | 0.022 | 0.3117 | No |
| 119 | PRF1 | NA | 15248 | 0.021 | 0.3092 | No |
| 120 | PEAR1 | NA | 15617 | 0.018 | 0.3029 | No |
| 121 | CCT7 | NA | 15831 | 0.017 | 0.2994 | No |
| 122 | KCNU1 | NA | 16058 | 0.016 | 0.2956 | No |
| 123 | IGHA1 | NA | 16252 | 0.015 | 0.2925 | No |
| 124 | CRISP1 | NA | 16256 | 0.015 | 0.2928 | No |
| 125 | CXCR4 | NA | 16294 | 0.014 | 0.2925 | No |
| 126 | NPTN | NA | 16346 | 0.014 | 0.2919 | No |
| 127 | APP | NA | 16779 | 0.012 | 0.2843 | No |
| 128 | RTN4 | NA | 18171 | 0.006 | 0.2588 | No |
| 129 | FCN1 | NA | 18571 | 0.004 | 0.2516 | No |
| 130 | ZP4 | NA | 18654 | 0.003 | 0.2502 | No |
| 131 | CADM1 | NA | 18908 | 0.002 | 0.2456 | No |
| 132 | CCT2 | NA | 25326 | -0.001 | 0.1276 | No |
| 133 | TMPRSS12 | NA | 25547 | -0.004 | 0.1236 | No |
| 134 | CASP3 | NA | 25593 | -0.004 | 0.1229 | No |
| 135 | LY6K | NA | 25605 | -0.005 | 0.1228 | No |
| 136 | PCDHB6 | NA | 25675 | -0.005 | 0.1217 | No |
| 137 | CRTAM | NA | 25811 | -0.007 | 0.1194 | No |
| 138 | IGHA2 | NA | 25884 | -0.007 | 0.1182 | No |
| 139 | DSCAM | NA | 26970 | -0.018 | 0.0987 | No |
| 140 | EMB | NA | 28137 | -0.029 | 0.0780 | No |
| 141 | FCN2 | NA | 28546 | -0.032 | 0.0714 | No |
| 142 | TULP1 | NA | 28612 | -0.033 | 0.0710 | No |
| 143 | VSTM2L | NA | 29569 | -0.042 | 0.0545 | No |
| 144 | MSN | NA | 29840 | -0.045 | 0.0507 | No |
| 145 | SEMA3A | NA | 30265 | -0.049 | 0.0442 | No |
| 146 | IZUMO1 | NA | 30468 | -0.051 | 0.0418 | No |
| 147 | FOXG1 | NA | 30809 | -0.055 | 0.0370 | No |
| 148 | B4GALT1 | NA | 31388 | -0.061 | 0.0279 | No |
| 149 | VCAN | NA | 31939 | -0.066 | 0.0195 | No |
| 150 | CNTNAP3 | NA | 32815 | -0.075 | 0.0053 | No |
| 151 | ADAM2 | NA | 33641 | -0.083 | -0.0077 | No |
| 152 | ARHGAP35 | NA | 33666 | -0.083 | -0.0060 | No |
| 153 | CNTN2 | NA | 33842 | -0.084 | -0.0071 | No |
| 154 | SPAM1 | NA | 33909 | -0.085 | -0.0061 | No |
| 155 | MYO18A | NA | 34105 | -0.087 | -0.0074 | No |
| 156 | HSPA1L | NA | 35030 | -0.097 | -0.0219 | No |
| 157 | ADAM30 | NA | 35194 | -0.098 | -0.0224 | No |
| 158 | PRSS37 | NA | 35292 | -0.099 | -0.0216 | No |
| 159 | PLA2G5 | NA | 35339 | -0.099 | -0.0199 | No |
| 160 | LGALS3 | NA | 35548 | -0.102 | -0.0211 | No |
| 161 | ZP3 | NA | 36824 | -0.114 | -0.0416 | No |
| 162 | CCT3 | NA | 37076 | -0.116 | -0.0433 | No |
| 163 | CATSPERD | NA | 37129 | -0.117 | -0.0412 | No |
| 164 | IZUMO1R | NA | 37323 | -0.119 | -0.0417 | No |
| 165 | NCK2 | NA | 37597 | -0.122 | -0.0436 | No |
| 166 | ASTL | NA | 37868 | -0.125 | -0.0453 | No |
| 167 | MBL2 | NA | 38004 | -0.126 | -0.0445 | No |
| 168 | DSCAML1 | NA | 38083 | -0.127 | -0.0427 | No |
| 169 | EXT1 | NA | 38666 | -0.134 | -0.0499 | No |
| 170 | MEGF10 | NA | 38681 | -0.134 | -0.0468 | No |
| 171 | VDAC2 | NA | 38788 | -0.135 | -0.0452 | No |
| 172 | ADAM5 | NA | 38873 | -0.136 | -0.0433 | No |
| 173 | PTX3 | NA | 39070 | -0.138 | -0.0433 | No |
| 174 | TEX101 | NA | 39282 | -0.140 | -0.0436 | No |
| 175 | ADAM32 | NA | 39862 | -0.147 | -0.0504 | No |
| 176 | NRCAM | NA | 40072 | -0.150 | -0.0504 | No |
| 177 | CD9 | NA | 40224 | -0.151 | -0.0493 | No |
| 178 | CCT8 | NA | 40352 | -0.153 | -0.0477 | No |
| 179 | DLG1 | NA | 40681 | -0.157 | -0.0497 | No |
| 180 | ADAM20 | NA | 40866 | -0.159 | -0.0490 | No |
| 181 | FEZF2 | NA | 41414 | -0.165 | -0.0548 | No |
| 182 | CNTN4 | NA | 41607 | -0.168 | -0.0539 | No |
| 183 | EPHB3 | NA | 41821 | -0.171 | -0.0535 | No |
| 184 | CCL19 | NA | 41896 | -0.172 | -0.0504 | No |
| 185 | ADAM21 | NA | 42457 | -0.179 | -0.0561 | No |
| 186 | LBP | NA | 42622 | -0.181 | -0.0544 | No |
| 187 | GAP43 | NA | 43144 | -0.188 | -0.0591 | No |
| 188 | CATSPERG | NA | 43276 | -0.189 | -0.0567 | No |
| 189 | PCDHA7 | NA | 43436 | -0.192 | -0.0547 | No |
| 190 | AMIGO1 | NA | 43490 | -0.192 | -0.0507 | No |
| 191 | FUT3 | NA | 43850 | -0.197 | -0.0522 | No |
| 192 | CCL21 | NA | 44349 | -0.205 | -0.0560 | No |
| 193 | NTM | NA | 44710 | -0.210 | -0.0572 | No |
| 194 | CLEC4M | NA | 45608 | -0.224 | -0.0680 | No |
| 195 | TNN | NA | 45638 | -0.224 | -0.0627 | No |
| 196 | NCAM2 | NA | 45818 | -0.227 | -0.0601 | No |
| 197 | PRSS55 | NA | 46213 | -0.234 | -0.0614 | No |
| 198 | UBAP2L | NA | 46262 | -0.234 | -0.0562 | No |
| 199 | FOLR1 | NA | 46336 | -0.236 | -0.0515 | No |
| 200 | ROBO1 | NA | 46341 | -0.236 | -0.0455 | No |
| 201 | ST6GALNAC6 | NA | 47156 | -0.250 | -0.0540 | No |
| 202 | ZPBP2 | NA | 47163 | -0.250 | -0.0476 | No |
| 203 | IGSF9 | NA | 47304 | -0.252 | -0.0437 | No |
| 204 | FOLR3 | NA | 48069 | -0.267 | -0.0509 | No |
| 205 | GLIPR1L2 | NA | 48116 | -0.268 | -0.0448 | No |
| 206 | CATSPER3 | NA | 48152 | -0.268 | -0.0386 | No |
| 207 | POMZP3 | NA | 48402 | -0.273 | -0.0361 | No |
| 208 | MEGF8 | NA | 48513 | -0.276 | -0.0310 | No |
| 209 | SPACA3 | NA | 49300 | -0.293 | -0.0379 | No |
| 210 | CATSPER2 | NA | 49541 | -0.299 | -0.0346 | No |
| 211 | CCT4 | NA | 49674 | -0.302 | -0.0292 | No |
| 212 | CD81 | NA | 49796 | -0.305 | -0.0236 | No |
| 213 | SCARB1 | NA | 49835 | -0.306 | -0.0164 | No |
| 214 | CNTN6 | NA | 49926 | -0.308 | -0.0102 | No |
| 215 | ROBO3 | NA | 50374 | -0.319 | -0.0101 | No |
| 216 | BSG | NA | 50627 | -0.326 | -0.0064 | No |
| 217 | OVGP1 | NA | 50632 | -0.326 | 0.0020 | No |
| 218 | TCP1 | NA | 51535 | -0.353 | -0.0055 | No |
| 219 | IGHE | NA | 51792 | -0.361 | -0.0009 | No |
| 220 | JMJD6 | NA | 52080 | -0.371 | 0.0034 | No |
| 221 | ADGRB1 | NA | 52214 | -0.377 | 0.0106 | No |
| 222 | PCSK4 | NA | 53006 | -0.411 | 0.0067 | No |
| 223 | CATSPER4 | NA | 53311 | -0.429 | 0.0122 | No |
| 224 | TUB | NA | 53470 | -0.440 | 0.0206 | No |