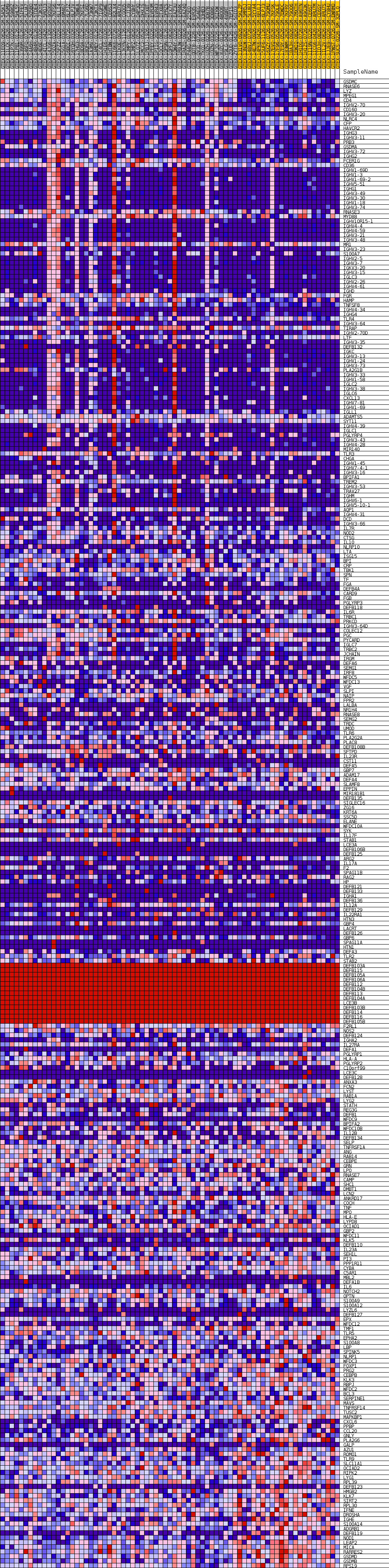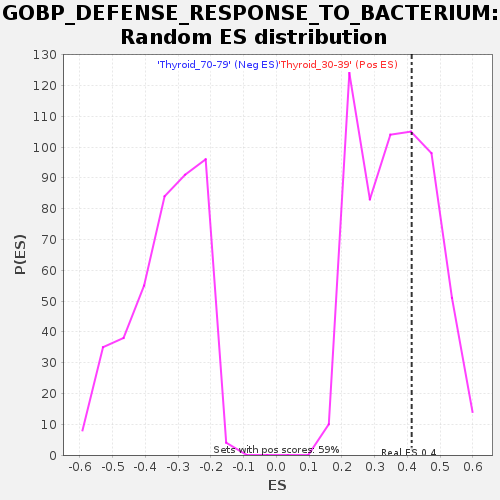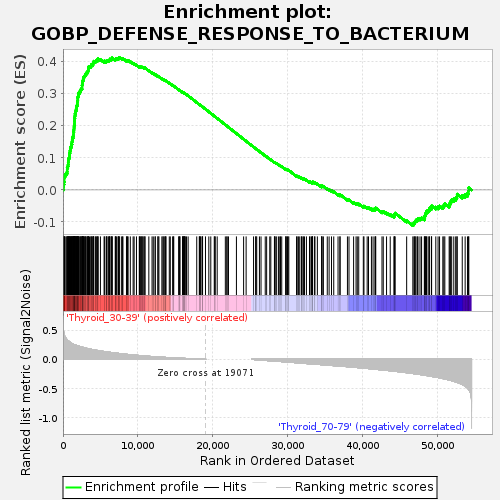
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Thyroid.Thyroid.cls #Thyroid_30-39_versus_Thyroid_70-79.Thyroid.cls #Thyroid_30-39_versus_Thyroid_70-79_repos |
| Phenotype | Thyroid.cls#Thyroid_30-39_versus_Thyroid_70-79_repos |
| Upregulated in class | Thyroid_30-39 |
| GeneSet | GOBP_DEFENSE_RESPONSE_TO_BACTERIUM |
| Enrichment Score (ES) | 0.41305098 |
| Normalized Enrichment Score (NES) | 1.1335667 |
| Nominal p-value | 0.37521222 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | GSDMC | NA | 59 | 0.493 | 0.0088 | Yes |
| 2 | RNASE6 | NA | 92 | 0.466 | 0.0176 | Yes |
| 3 | LYZ | NA | 157 | 0.424 | 0.0250 | Yes |
| 4 | MPEG1 | NA | 180 | 0.415 | 0.0329 | Yes |
| 5 | CD4 | NA | 181 | 0.413 | 0.0412 | Yes |
| 6 | IGHV2-70 | NA | 266 | 0.384 | 0.0474 | Yes |
| 7 | CD160 | NA | 413 | 0.354 | 0.0519 | Yes |
| 8 | IGHV3-20 | NA | 541 | 0.333 | 0.0562 | Yes |
| 9 | NLRC4 | NA | 558 | 0.331 | 0.0626 | Yes |
| 10 | CFP | NA | 562 | 0.330 | 0.0692 | Yes |
| 11 | HAVCR2 | NA | 641 | 0.321 | 0.0742 | Yes |
| 12 | IGHG3 | NA | 685 | 0.317 | 0.0798 | Yes |
| 13 | IGHV3-11 | NA | 702 | 0.315 | 0.0859 | Yes |
| 14 | PRB3 | NA | 719 | 0.313 | 0.0919 | Yes |
| 15 | GSDMA | NA | 739 | 0.311 | 0.0978 | Yes |
| 16 | IGHV3-72 | NA | 821 | 0.303 | 0.1024 | Yes |
| 17 | IGHG2 | NA | 846 | 0.301 | 0.1080 | Yes |
| 18 | FCER1G | NA | 868 | 0.298 | 0.1136 | Yes |
| 19 | CD36 | NA | 926 | 0.292 | 0.1185 | Yes |
| 20 | IGHV1-69D | NA | 947 | 0.290 | 0.1239 | Yes |
| 21 | IGHV1-3 | NA | 1000 | 0.286 | 0.1287 | Yes |
| 22 | IGHV1-69-2 | NA | 1014 | 0.284 | 0.1342 | Yes |
| 23 | IGHV5-51 | NA | 1135 | 0.274 | 0.1375 | Yes |
| 24 | IGHG1 | NA | 1146 | 0.273 | 0.1429 | Yes |
| 25 | IGHV3-49 | NA | 1199 | 0.270 | 0.1473 | Yes |
| 26 | IGHV3-30 | NA | 1213 | 0.269 | 0.1525 | Yes |
| 27 | IGHV1-18 | NA | 1236 | 0.267 | 0.1575 | Yes |
| 28 | IGHV3-74 | NA | 1243 | 0.267 | 0.1627 | Yes |
| 29 | RNASE3 | NA | 1348 | 0.260 | 0.1661 | Yes |
| 30 | MYD88 | NA | 1367 | 0.259 | 0.1709 | Yes |
| 31 | IGHV1OR15-1 | NA | 1378 | 0.258 | 0.1760 | Yes |
| 32 | IGHV4-4 | NA | 1379 | 0.258 | 0.1811 | Yes |
| 33 | IGHV4-59 | NA | 1451 | 0.254 | 0.1850 | Yes |
| 34 | IGHV3-21 | NA | 1460 | 0.253 | 0.1899 | Yes |
| 35 | IGHV3-48 | NA | 1478 | 0.252 | 0.1947 | Yes |
| 36 | MR1 | NA | 1486 | 0.252 | 0.1996 | Yes |
| 37 | IGHV3-23 | NA | 1510 | 0.251 | 0.2042 | Yes |
| 38 | S100A7 | NA | 1533 | 0.249 | 0.2089 | Yes |
| 39 | IGHV2-5 | NA | 1536 | 0.249 | 0.2138 | Yes |
| 40 | IGHV3-7 | NA | 1550 | 0.248 | 0.2186 | Yes |
| 41 | IGKV3-20 | NA | 1555 | 0.248 | 0.2235 | Yes |
| 42 | IGHV3-15 | NA | 1586 | 0.246 | 0.2279 | Yes |
| 43 | IGLC3 | NA | 1602 | 0.246 | 0.2326 | Yes |
| 44 | IGHV2-26 | NA | 1612 | 0.245 | 0.2374 | Yes |
| 45 | IGHV4-61 | NA | 1690 | 0.242 | 0.2408 | Yes |
| 46 | IGHD | NA | 1697 | 0.241 | 0.2456 | Yes |
| 47 | FGR | NA | 1741 | 0.239 | 0.2496 | Yes |
| 48 | HAMP | NA | 1771 | 0.237 | 0.2538 | Yes |
| 49 | TNFSF8 | NA | 1788 | 0.237 | 0.2583 | Yes |
| 50 | IGHV4-34 | NA | 1818 | 0.235 | 0.2625 | Yes |
| 51 | IGHG4 | NA | 1903 | 0.232 | 0.2656 | Yes |
| 52 | TLR4 | NA | 1930 | 0.231 | 0.2698 | Yes |
| 53 | IGHV3-64 | NA | 1932 | 0.231 | 0.2744 | Yes |
| 54 | TIRAP | NA | 1967 | 0.229 | 0.2784 | Yes |
| 55 | IGHV2-70D | NA | 1979 | 0.229 | 0.2828 | Yes |
| 56 | LTF | NA | 1983 | 0.229 | 0.2874 | Yes |
| 57 | IGHV3-35 | NA | 2011 | 0.228 | 0.2915 | Yes |
| 58 | DEFB132 | NA | 2045 | 0.227 | 0.2954 | Yes |
| 59 | IGKC | NA | 2057 | 0.226 | 0.2998 | Yes |
| 60 | IGHV3-13 | NA | 2110 | 0.224 | 0.3033 | Yes |
| 61 | IGHV1-24 | NA | 2244 | 0.219 | 0.3053 | Yes |
| 62 | IGHV3-73 | NA | 2283 | 0.217 | 0.3090 | Yes |
| 63 | PLA2G1B | NA | 2366 | 0.214 | 0.3118 | Yes |
| 64 | IGHV3-33 | NA | 2419 | 0.212 | 0.3151 | Yes |
| 65 | IGHV1-58 | NA | 2539 | 0.208 | 0.3171 | Yes |
| 66 | IGLC2 | NA | 2547 | 0.208 | 0.3211 | Yes |
| 67 | IGHV3-38 | NA | 2548 | 0.208 | 0.3253 | Yes |
| 68 | IGLC6 | NA | 2574 | 0.207 | 0.3290 | Yes |
| 69 | CXCL13 | NA | 2612 | 0.205 | 0.3325 | Yes |
| 70 | IGHV7-81 | NA | 2622 | 0.205 | 0.3364 | Yes |
| 71 | IGHV1-69 | NA | 2627 | 0.205 | 0.3405 | Yes |
| 72 | IGLL1 | NA | 2697 | 0.202 | 0.3433 | Yes |
| 73 | ADAMTS5 | NA | 2710 | 0.202 | 0.3471 | Yes |
| 74 | SYT11 | NA | 2726 | 0.201 | 0.3509 | Yes |
| 75 | IGHV4-39 | NA | 2816 | 0.199 | 0.3533 | Yes |
| 76 | IGLC1 | NA | 2899 | 0.195 | 0.3557 | Yes |
| 77 | PGLYRP4 | NA | 2962 | 0.193 | 0.3584 | Yes |
| 78 | IGHV3-43 | NA | 3029 | 0.191 | 0.3611 | Yes |
| 79 | IGHV4-28 | NA | 3075 | 0.190 | 0.3641 | Yes |
| 80 | MIR140 | NA | 3211 | 0.186 | 0.3653 | Yes |
| 81 | TLR3 | NA | 3286 | 0.184 | 0.3677 | Yes |
| 82 | CHGA | NA | 3323 | 0.183 | 0.3707 | Yes |
| 83 | IGHV1-45 | NA | 3364 | 0.182 | 0.3736 | Yes |
| 84 | IGHV7-4-1 | NA | 3393 | 0.181 | 0.3767 | Yes |
| 85 | IGHV3-16 | NA | 3394 | 0.181 | 0.3804 | Yes |
| 86 | BPIFA1 | NA | 3403 | 0.181 | 0.3839 | Yes |
| 87 | TREM2 | NA | 3542 | 0.176 | 0.3849 | Yes |
| 88 | IGHV3-53 | NA | 3723 | 0.172 | 0.3850 | Yes |
| 89 | TRAV27 | NA | 3761 | 0.171 | 0.3878 | Yes |
| 90 | IGHM | NA | 3785 | 0.170 | 0.3907 | Yes |
| 91 | IGHV6-1 | NA | 3991 | 0.165 | 0.3903 | Yes |
| 92 | IGHV5-10-1 | NA | 4000 | 0.164 | 0.3934 | Yes |
| 93 | AQP1 | NA | 4034 | 0.163 | 0.3961 | Yes |
| 94 | IGHV4-31 | NA | 4069 | 0.163 | 0.3988 | Yes |
| 95 | DCD | NA | 4115 | 0.161 | 0.4012 | Yes |
| 96 | IGHV3-66 | NA | 4327 | 0.156 | 0.4005 | Yes |
| 97 | IL7R | NA | 4403 | 0.155 | 0.4022 | Yes |
| 98 | NOD2 | NA | 4498 | 0.152 | 0.4035 | Yes |
| 99 | CTSG | NA | 4631 | 0.149 | 0.4041 | Yes |
| 100 | IL10 | NA | 4670 | 0.149 | 0.4064 | Yes |
| 101 | NLRP10 | NA | 4673 | 0.149 | 0.4093 | Yes |
| 102 | LTA | NA | 5001 | 0.142 | 0.4062 | Yes |
| 103 | ISG15 | NA | 5512 | 0.132 | 0.3994 | Yes |
| 104 | BPI | NA | 5515 | 0.132 | 0.4021 | Yes |
| 105 | CRP | NA | 5763 | 0.127 | 0.4001 | Yes |
| 106 | TBK1 | NA | 5833 | 0.126 | 0.4013 | Yes |
| 107 | SPN | NA | 5840 | 0.126 | 0.4038 | Yes |
| 108 | TF | NA | 6027 | 0.123 | 0.4028 | Yes |
| 109 | FGA | NA | 6108 | 0.121 | 0.4038 | Yes |
| 110 | DEFB4A | NA | 6209 | 0.120 | 0.4043 | Yes |
| 111 | CARD9 | NA | 6229 | 0.119 | 0.4064 | Yes |
| 112 | FGB | NA | 6275 | 0.118 | 0.4079 | Yes |
| 113 | PGLYRP3 | NA | 6455 | 0.116 | 0.4070 | Yes |
| 114 | DEFB118 | NA | 6515 | 0.114 | 0.4082 | Yes |
| 115 | IL6R | NA | 6557 | 0.114 | 0.4097 | Yes |
| 116 | TRBC1 | NA | 6565 | 0.114 | 0.4119 | Yes |
| 117 | PRKCD | NA | 6985 | 0.107 | 0.4063 | Yes |
| 118 | IGHV3-64D | NA | 7005 | 0.107 | 0.4081 | Yes |
| 119 | COLEC12 | NA | 7051 | 0.106 | 0.4094 | Yes |
| 120 | PGC | NA | 7162 | 0.104 | 0.4095 | Yes |
| 121 | PYCARD | NA | 7352 | 0.101 | 0.4081 | Yes |
| 122 | IGLC7 | NA | 7419 | 0.100 | 0.4089 | Yes |
| 123 | TRBC2 | NA | 7436 | 0.100 | 0.4106 | Yes |
| 124 | JCHAIN | NA | 7504 | 0.099 | 0.4113 | Yes |
| 125 | IRGM | NA | 7519 | 0.098 | 0.4131 | Yes |
| 126 | DEFA6 | NA | 7799 | 0.095 | 0.4098 | No |
| 127 | SEMG1 | NA | 7962 | 0.092 | 0.4087 | No |
| 128 | IRF8 | NA | 8001 | 0.092 | 0.4098 | No |
| 129 | WFDC5 | NA | 8480 | 0.085 | 0.4027 | No |
| 130 | WFDC13 | NA | 8526 | 0.084 | 0.4036 | No |
| 131 | VGF | NA | 8613 | 0.083 | 0.4037 | No |
| 132 | SLPI | NA | 8698 | 0.082 | 0.4038 | No |
| 133 | NAIP | NA | 8994 | 0.078 | 0.3999 | No |
| 134 | FPR2 | NA | 9369 | 0.074 | 0.3945 | No |
| 135 | LALBA | NA | 9502 | 0.072 | 0.3935 | No |
| 136 | NR1H4 | NA | 9816 | 0.068 | 0.3891 | No |
| 137 | RNASE8 | NA | 10194 | 0.064 | 0.3835 | No |
| 138 | SEMG2 | NA | 10218 | 0.064 | 0.3843 | No |
| 139 | TRDC | NA | 10333 | 0.062 | 0.3835 | No |
| 140 | UMOD | NA | 10418 | 0.061 | 0.3832 | No |
| 141 | TLR6 | NA | 10518 | 0.060 | 0.3826 | No |
| 142 | PLA2G2A | NA | 10572 | 0.060 | 0.3828 | No |
| 143 | PLAC8 | NA | 10674 | 0.059 | 0.3821 | No |
| 144 | DEFB108B | NA | 10805 | 0.057 | 0.3809 | No |
| 145 | SFTPD | NA | 10942 | 0.056 | 0.3795 | No |
| 146 | IL23R | NA | 10943 | 0.056 | 0.3806 | No |
| 147 | CST11 | NA | 11468 | 0.050 | 0.3720 | No |
| 148 | DEFA5 | NA | 11898 | 0.046 | 0.3650 | No |
| 149 | GBP7 | NA | 12061 | 0.045 | 0.3629 | No |
| 150 | ADAM17 | NA | 12310 | 0.042 | 0.3592 | No |
| 151 | DEFA4 | NA | 12325 | 0.042 | 0.3598 | No |
| 152 | SLAMF8 | NA | 12688 | 0.039 | 0.3539 | No |
| 153 | EPPIN | NA | 12690 | 0.039 | 0.3547 | No |
| 154 | MIR181B1 | NA | 12815 | 0.038 | 0.3532 | No |
| 155 | DEFB135 | NA | 13211 | 0.035 | 0.3466 | No |
| 156 | SIGLEC16 | NA | 13367 | 0.034 | 0.3444 | No |
| 157 | ZG16 | NA | 13391 | 0.034 | 0.3447 | No |
| 158 | KRT6A | NA | 13606 | 0.032 | 0.3414 | No |
| 159 | SSC5D | NA | 13642 | 0.032 | 0.3414 | No |
| 160 | ELANE | NA | 13717 | 0.031 | 0.3406 | No |
| 161 | WFDC10A | NA | 13757 | 0.031 | 0.3405 | No |
| 162 | SYK | NA | 14202 | 0.028 | 0.3329 | No |
| 163 | IL17F | NA | 14335 | 0.027 | 0.3310 | No |
| 164 | STAB1 | NA | 14649 | 0.024 | 0.3257 | No |
| 165 | LCE3A | NA | 14745 | 0.024 | 0.3245 | No |
| 166 | DEFB106B | NA | 14798 | 0.024 | 0.3240 | No |
| 167 | DEFB125 | NA | 15456 | 0.019 | 0.3123 | No |
| 168 | ARG2 | NA | 15513 | 0.019 | 0.3116 | No |
| 169 | IL17A | NA | 15583 | 0.018 | 0.3107 | No |
| 170 | F2 | NA | 15633 | 0.018 | 0.3102 | No |
| 171 | SPAG11B | NA | 15971 | 0.016 | 0.3043 | No |
| 172 | RAG2 | NA | 16005 | 0.016 | 0.3040 | No |
| 173 | HP | NA | 16048 | 0.016 | 0.3035 | No |
| 174 | DEFB121 | NA | 16077 | 0.015 | 0.3033 | No |
| 175 | DEFB133 | NA | 16114 | 0.015 | 0.3030 | No |
| 176 | IGHA1 | NA | 16252 | 0.015 | 0.3007 | No |
| 177 | DEFB136 | NA | 16255 | 0.015 | 0.3010 | No |
| 178 | IL12A | NA | 16263 | 0.014 | 0.3011 | No |
| 179 | DEFB129 | NA | 16333 | 0.014 | 0.3002 | No |
| 180 | IL22RA1 | NA | 16499 | 0.013 | 0.2974 | No |
| 181 | HTN3 | NA | 16717 | 0.012 | 0.2936 | No |
| 182 | GBP4 | NA | 17869 | 0.007 | 0.2726 | No |
| 183 | LACRT | NA | 18210 | 0.005 | 0.2664 | No |
| 184 | DEFB126 | NA | 18240 | 0.005 | 0.2660 | No |
| 185 | GBP6 | NA | 18332 | 0.005 | 0.2644 | No |
| 186 | SPAG11A | NA | 18351 | 0.005 | 0.2642 | No |
| 187 | HTN1 | NA | 18582 | 0.004 | 0.2600 | No |
| 188 | DEFA3 | NA | 18608 | 0.004 | 0.2596 | No |
| 189 | TLR2 | NA | 18649 | 0.003 | 0.2589 | No |
| 190 | STAB2 | NA | 19037 | 0.000 | 0.2518 | No |
| 191 | DEFB103A | NA | 19484 | 0.000 | 0.2436 | No |
| 192 | DEFB115 | NA | 19747 | 0.000 | 0.2388 | No |
| 193 | DEFB105A | NA | 20210 | 0.000 | 0.2303 | No |
| 194 | DEFB106A | NA | 20276 | 0.000 | 0.2291 | No |
| 195 | DEFB112 | NA | 20339 | 0.000 | 0.2279 | No |
| 196 | DEFB104B | NA | 20602 | 0.000 | 0.2231 | No |
| 197 | DEFB113 | NA | 21689 | 0.000 | 0.2031 | No |
| 198 | DEFB104A | NA | 21882 | 0.000 | 0.1996 | No |
| 199 | LCE3B | NA | 21934 | 0.000 | 0.1986 | No |
| 200 | DEFB103B | NA | 22111 | 0.000 | 0.1954 | No |
| 201 | DEFB114 | NA | 23172 | 0.000 | 0.1758 | No |
| 202 | DEFB116 | NA | 24132 | 0.000 | 0.1582 | No |
| 203 | DEFB105B | NA | 24468 | 0.000 | 0.1520 | No |
| 204 | F2RL1 | NA | 25460 | -0.003 | 0.1338 | No |
| 205 | NOS2 | NA | 25752 | -0.006 | 0.1286 | No |
| 206 | DEFB124 | NA | 25767 | -0.006 | 0.1284 | No |
| 207 | IGHA2 | NA | 25884 | -0.007 | 0.1264 | No |
| 208 | IL27RA | NA | 26246 | -0.011 | 0.1200 | No |
| 209 | DEFA1 | NA | 26458 | -0.013 | 0.1164 | No |
| 210 | PGLYRP1 | NA | 27022 | -0.018 | 0.1064 | No |
| 211 | HLA-A | NA | 27209 | -0.020 | 0.1034 | No |
| 212 | PGLYRP2 | NA | 27626 | -0.024 | 0.0962 | No |
| 213 | C10orf99 | NA | 27767 | -0.025 | 0.0941 | No |
| 214 | LCE3C | NA | 28250 | -0.030 | 0.0858 | No |
| 215 | DEFB128 | NA | 28284 | -0.030 | 0.0858 | No |
| 216 | ANXA3 | NA | 28351 | -0.031 | 0.0852 | No |
| 217 | FCN2 | NA | 28546 | -0.032 | 0.0823 | No |
| 218 | LYST | NA | 28803 | -0.035 | 0.0783 | No |
| 219 | RAB1A | NA | 28966 | -0.036 | 0.0760 | No |
| 220 | LYG2 | NA | 29063 | -0.037 | 0.0750 | No |
| 221 | STATH | NA | 29227 | -0.039 | 0.0728 | No |
| 222 | REG3G | NA | 29761 | -0.044 | 0.0639 | No |
| 223 | DEFB1 | NA | 29780 | -0.045 | 0.0644 | No |
| 224 | WFDC9 | NA | 29831 | -0.045 | 0.0644 | No |
| 225 | BPIFA2 | NA | 29993 | -0.047 | 0.0624 | No |
| 226 | WFDC10B | NA | 30069 | -0.048 | 0.0620 | No |
| 227 | IL12B | NA | 30171 | -0.049 | 0.0611 | No |
| 228 | DEFB134 | NA | 31203 | -0.059 | 0.0433 | No |
| 229 | SELP | NA | 31285 | -0.060 | 0.0430 | No |
| 230 | TNFRSF1A | NA | 31450 | -0.061 | 0.0412 | No |
| 231 | ANG | NA | 31568 | -0.062 | 0.0403 | No |
| 232 | RAB14 | NA | 31828 | -0.065 | 0.0368 | No |
| 233 | CEBPE | NA | 31862 | -0.065 | 0.0375 | No |
| 234 | GRN | NA | 32056 | -0.067 | 0.0353 | No |
| 235 | LPO | NA | 32114 | -0.068 | 0.0356 | No |
| 236 | RNASE7 | NA | 32270 | -0.069 | 0.0342 | No |
| 237 | CAMP | NA | 32525 | -0.072 | 0.0309 | No |
| 238 | SHC1 | NA | 32934 | -0.076 | 0.0250 | No |
| 239 | DMBT1 | NA | 33009 | -0.077 | 0.0251 | No |
| 240 | LCN2 | NA | 33202 | -0.079 | 0.0232 | No |
| 241 | ANKRD17 | NA | 33314 | -0.080 | 0.0228 | No |
| 242 | COCH | NA | 33325 | -0.080 | 0.0242 | No |
| 243 | TNF | NA | 33344 | -0.080 | 0.0254 | No |
| 244 | MPO | NA | 33594 | -0.082 | 0.0225 | No |
| 245 | HLA-E | NA | 33665 | -0.083 | 0.0229 | No |
| 246 | LYPD8 | NA | 33986 | -0.086 | 0.0187 | No |
| 247 | OCIAD1 | NA | 34550 | -0.092 | 0.0102 | No |
| 248 | GBP2 | NA | 34581 | -0.092 | 0.0115 | No |
| 249 | WFDC11 | NA | 34610 | -0.092 | 0.0128 | No |
| 250 | KLK5 | NA | 34791 | -0.094 | 0.0114 | No |
| 251 | DEFB110 | NA | 35310 | -0.099 | 0.0039 | No |
| 252 | IL23A | NA | 35556 | -0.102 | 0.0014 | No |
| 253 | SEH1L | NA | 35875 | -0.105 | -0.0024 | No |
| 254 | PI3 | NA | 36174 | -0.108 | -0.0057 | No |
| 255 | PPP1R11 | NA | 36749 | -0.113 | -0.0140 | No |
| 256 | CYBA | NA | 37003 | -0.116 | -0.0163 | No |
| 257 | C5AR1 | NA | 37017 | -0.116 | -0.0142 | No |
| 258 | MBL2 | NA | 38004 | -0.126 | -0.0298 | No |
| 259 | DEFA1B | NA | 38212 | -0.129 | -0.0311 | No |
| 260 | IL6 | NA | 38829 | -0.135 | -0.0397 | No |
| 261 | NOTCH2 | NA | 39157 | -0.139 | -0.0429 | No |
| 262 | OPTN | NA | 39319 | -0.141 | -0.0430 | No |
| 263 | S100A9 | NA | 39514 | -0.143 | -0.0437 | No |
| 264 | S100A12 | NA | 40126 | -0.150 | -0.0520 | No |
| 265 | LYZL6 | NA | 40212 | -0.151 | -0.0505 | No |
| 266 | DEFB127 | NA | 40641 | -0.156 | -0.0552 | No |
| 267 | EPX | NA | 40819 | -0.158 | -0.0553 | No |
| 268 | WFDC12 | NA | 41210 | -0.163 | -0.0592 | No |
| 269 | TMF1 | NA | 41427 | -0.166 | -0.0599 | No |
| 270 | TLR5 | NA | 41679 | -0.169 | -0.0611 | No |
| 271 | EPHA2 | NA | 41746 | -0.170 | -0.0589 | No |
| 272 | S100A8 | NA | 41788 | -0.170 | -0.0562 | No |
| 273 | LBP | NA | 42622 | -0.181 | -0.0679 | No |
| 274 | SPINK5 | NA | 42789 | -0.183 | -0.0673 | No |
| 275 | NLRP1 | NA | 43231 | -0.189 | -0.0716 | No |
| 276 | WFDC3 | NA | 43722 | -0.196 | -0.0767 | No |
| 277 | FOXP1 | NA | 44223 | -0.203 | -0.0818 | No |
| 278 | PRG2 | NA | 44269 | -0.204 | -0.0785 | No |
| 279 | CEBPB | NA | 44297 | -0.204 | -0.0749 | No |
| 280 | KLK3 | NA | 44383 | -0.206 | -0.0723 | No |
| 281 | RBPJ | NA | 45918 | -0.229 | -0.0960 | No |
| 282 | WFDC2 | NA | 46744 | -0.243 | -0.1063 | No |
| 283 | BCL3 | NA | 46892 | -0.245 | -0.1041 | No |
| 284 | SERPINE1 | NA | 46977 | -0.247 | -0.1007 | No |
| 285 | MAVS | NA | 47058 | -0.249 | -0.0971 | No |
| 286 | TNFRSF14 | NA | 47166 | -0.250 | -0.0941 | No |
| 287 | TUSC2 | NA | 47352 | -0.253 | -0.0924 | No |
| 288 | MAPKBP1 | NA | 47432 | -0.254 | -0.0887 | No |
| 289 | CXCL6 | NA | 47701 | -0.260 | -0.0884 | No |
| 290 | PPBP | NA | 47910 | -0.264 | -0.0869 | No |
| 291 | CCL20 | NA | 48280 | -0.271 | -0.0883 | No |
| 292 | GNLY | NA | 48334 | -0.272 | -0.0838 | No |
| 293 | PLA2G6 | NA | 48363 | -0.273 | -0.0788 | No |
| 294 | GALP | NA | 48450 | -0.275 | -0.0749 | No |
| 295 | AZU1 | NA | 48507 | -0.276 | -0.0704 | No |
| 296 | ROMO1 | NA | 48570 | -0.277 | -0.0659 | No |
| 297 | TLR9 | NA | 48781 | -0.282 | -0.0641 | No |
| 298 | SLC11A1 | NA | 48922 | -0.285 | -0.0610 | No |
| 299 | OCIAD2 | NA | 48984 | -0.286 | -0.0563 | No |
| 300 | RIPK2 | NA | 49236 | -0.292 | -0.0551 | No |
| 301 | LYG1 | NA | 49252 | -0.292 | -0.0495 | No |
| 302 | RPL39 | NA | 49811 | -0.305 | -0.0536 | No |
| 303 | DEFB123 | NA | 50115 | -0.312 | -0.0529 | No |
| 304 | HMGB2 | NA | 50313 | -0.317 | -0.0502 | No |
| 305 | KLK7 | NA | 50750 | -0.329 | -0.0516 | No |
| 306 | SIRT2 | NA | 50883 | -0.333 | -0.0473 | No |
| 307 | RPL30 | NA | 51017 | -0.337 | -0.0430 | No |
| 308 | IFNE | NA | 51596 | -0.354 | -0.0465 | No |
| 309 | DROSHA | NA | 51687 | -0.358 | -0.0409 | No |
| 310 | IGHE | NA | 51792 | -0.361 | -0.0356 | No |
| 311 | S100A14 | NA | 51933 | -0.366 | -0.0308 | No |
| 312 | ADGRB1 | NA | 52214 | -0.377 | -0.0284 | No |
| 313 | DEFB119 | NA | 52468 | -0.386 | -0.0253 | No |
| 314 | NOD1 | NA | 52624 | -0.392 | -0.0202 | No |
| 315 | LEAP2 | NA | 52673 | -0.394 | -0.0132 | No |
| 316 | MICA | NA | 53345 | -0.432 | -0.0169 | No |
| 317 | RARRES2 | NA | 53732 | -0.460 | -0.0147 | No |
| 318 | GSDMD | NA | 54044 | -0.490 | -0.0106 | No |
| 319 | GSDMB | NA | 54190 | -0.510 | -0.0030 | No |
| 320 | RPS19 | NA | 54218 | -0.515 | 0.0069 | No |