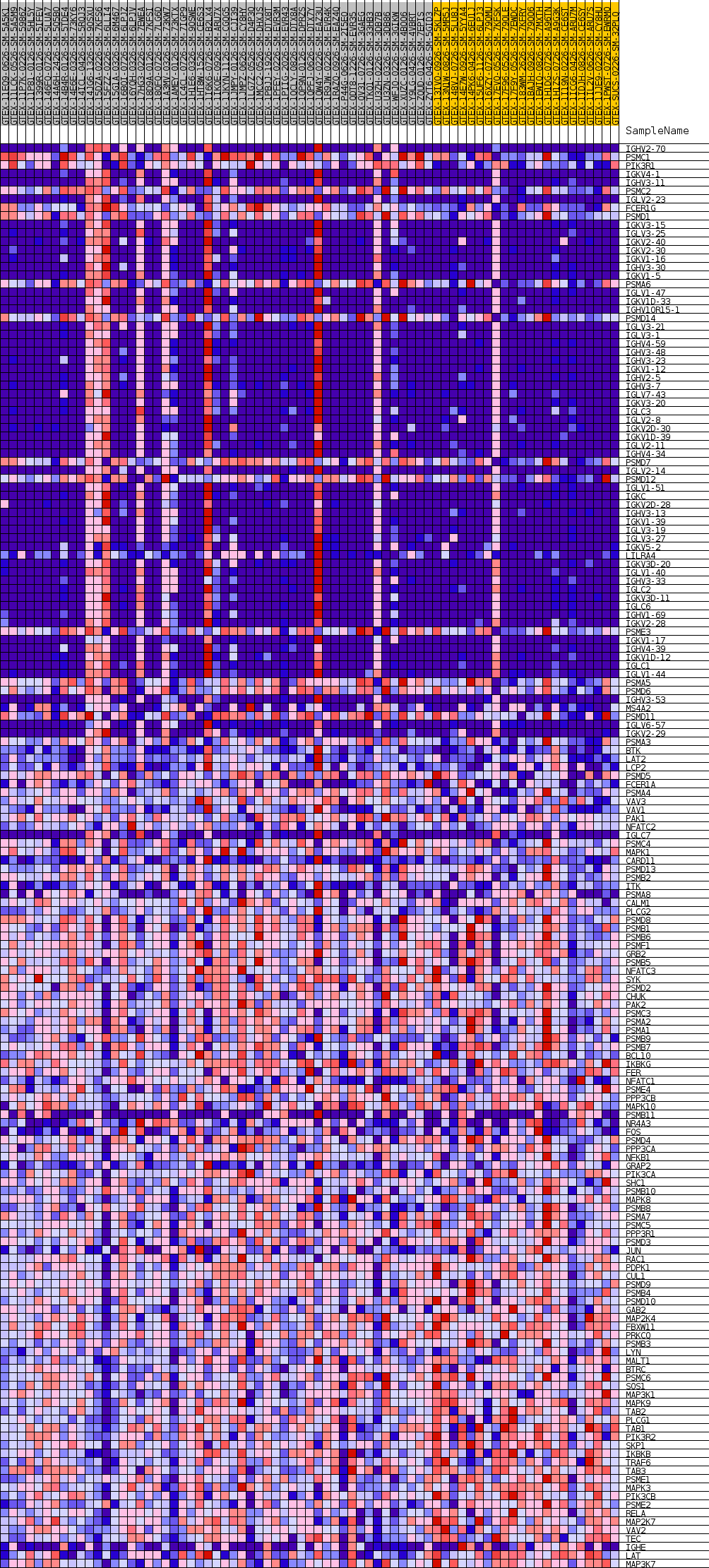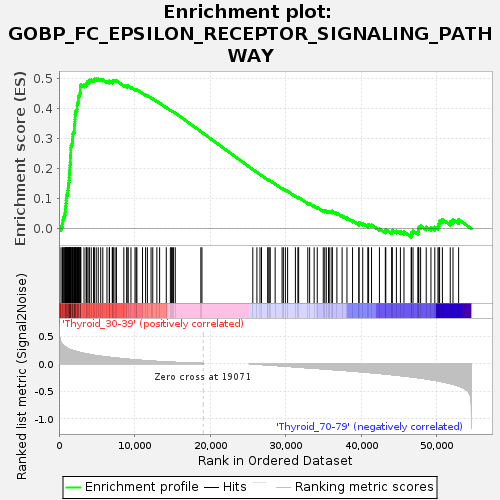
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Thyroid.Thyroid.cls #Thyroid_30-39_versus_Thyroid_70-79.Thyroid.cls #Thyroid_30-39_versus_Thyroid_70-79_repos |
| Phenotype | Thyroid.cls#Thyroid_30-39_versus_Thyroid_70-79_repos |
| Upregulated in class | Thyroid_30-39 |
| GeneSet | GOBP_FC_EPSILON_RECEPTOR_SIGNALING_PATHWAY |
| Enrichment Score (ES) | 0.4990286 |
| Normalized Enrichment Score (NES) | 1.1996613 |
| Nominal p-value | 0.30053666 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | IGHV2-70 | NA | 266 | 0.384 | 0.0083 | Yes |
| 2 | PSMC1 | NA | 442 | 0.349 | 0.0171 | Yes |
| 3 | PIK3R1 | NA | 488 | 0.341 | 0.0281 | Yes |
| 4 | IGKV4-1 | NA | 570 | 0.329 | 0.0379 | Yes |
| 5 | IGHV3-11 | NA | 702 | 0.315 | 0.0463 | Yes |
| 6 | PSMC2 | NA | 804 | 0.305 | 0.0550 | Yes |
| 7 | IGLV2-23 | NA | 825 | 0.303 | 0.0650 | Yes |
| 8 | FCER1G | NA | 868 | 0.298 | 0.0745 | Yes |
| 9 | PSMD1 | NA | 914 | 0.294 | 0.0838 | Yes |
| 10 | IGKV3-15 | NA | 938 | 0.291 | 0.0934 | Yes |
| 11 | IGLV3-25 | NA | 962 | 0.289 | 0.1029 | Yes |
| 12 | IGKV2-40 | NA | 1015 | 0.284 | 0.1118 | Yes |
| 13 | IGKV2-30 | NA | 1114 | 0.276 | 0.1194 | Yes |
| 14 | IGKV1-16 | NA | 1158 | 0.272 | 0.1280 | Yes |
| 15 | IGHV3-30 | NA | 1213 | 0.269 | 0.1363 | Yes |
| 16 | IGKV1-5 | NA | 1241 | 0.267 | 0.1450 | Yes |
| 17 | PSMA6 | NA | 1288 | 0.264 | 0.1532 | Yes |
| 18 | IGLV1-47 | NA | 1327 | 0.261 | 0.1615 | Yes |
| 19 | IGKV1D-33 | NA | 1341 | 0.260 | 0.1702 | Yes |
| 20 | IGHV1OR15-1 | NA | 1378 | 0.258 | 0.1785 | Yes |
| 21 | PSMD14 | NA | 1385 | 0.258 | 0.1872 | Yes |
| 22 | IGLV3-21 | NA | 1420 | 0.255 | 0.1954 | Yes |
| 23 | IGLV3-1 | NA | 1435 | 0.255 | 0.2039 | Yes |
| 24 | IGHV4-59 | NA | 1451 | 0.254 | 0.2124 | Yes |
| 25 | IGHV3-48 | NA | 1478 | 0.252 | 0.2206 | Yes |
| 26 | IGHV3-23 | NA | 1510 | 0.251 | 0.2286 | Yes |
| 27 | IGKV1-12 | NA | 1511 | 0.251 | 0.2373 | Yes |
| 28 | IGHV2-5 | NA | 1536 | 0.249 | 0.2454 | Yes |
| 29 | IGHV3-7 | NA | 1550 | 0.248 | 0.2537 | Yes |
| 30 | IGLV7-43 | NA | 1552 | 0.248 | 0.2622 | Yes |
| 31 | IGKV3-20 | NA | 1555 | 0.248 | 0.2707 | Yes |
| 32 | IGLC3 | NA | 1602 | 0.246 | 0.2784 | Yes |
| 33 | IGLV2-8 | NA | 1729 | 0.239 | 0.2843 | Yes |
| 34 | IGKV2D-30 | NA | 1768 | 0.238 | 0.2918 | Yes |
| 35 | IGKV1D-39 | NA | 1794 | 0.236 | 0.2994 | Yes |
| 36 | IGLV2-11 | NA | 1811 | 0.236 | 0.3073 | Yes |
| 37 | IGHV4-34 | NA | 1818 | 0.235 | 0.3153 | Yes |
| 38 | PSMD7 | NA | 1891 | 0.232 | 0.3219 | Yes |
| 39 | IGLV2-14 | NA | 2033 | 0.227 | 0.3272 | Yes |
| 40 | PSMD12 | NA | 2042 | 0.227 | 0.3348 | Yes |
| 41 | IGLV1-51 | NA | 2044 | 0.227 | 0.3426 | Yes |
| 42 | IGKC | NA | 2057 | 0.226 | 0.3502 | Yes |
| 43 | IGKV2D-28 | NA | 2099 | 0.225 | 0.3572 | Yes |
| 44 | IGHV3-13 | NA | 2110 | 0.224 | 0.3647 | Yes |
| 45 | IGKV1-39 | NA | 2126 | 0.224 | 0.3721 | Yes |
| 46 | IGLV3-19 | NA | 2132 | 0.224 | 0.3797 | Yes |
| 47 | IGLV3-27 | NA | 2209 | 0.221 | 0.3859 | Yes |
| 48 | IGKV5-2 | NA | 2231 | 0.220 | 0.3931 | Yes |
| 49 | LILRA4 | NA | 2371 | 0.214 | 0.3979 | Yes |
| 50 | IGKV3D-20 | NA | 2398 | 0.213 | 0.4048 | Yes |
| 51 | IGLV1-40 | NA | 2400 | 0.213 | 0.4121 | Yes |
| 52 | IGHV3-33 | NA | 2419 | 0.212 | 0.4191 | Yes |
| 53 | IGLC2 | NA | 2547 | 0.208 | 0.4239 | Yes |
| 54 | IGKV3D-11 | NA | 2573 | 0.207 | 0.4306 | Yes |
| 55 | IGLC6 | NA | 2574 | 0.207 | 0.4377 | Yes |
| 56 | IGHV1-69 | NA | 2627 | 0.205 | 0.4438 | Yes |
| 57 | IGKV2-28 | NA | 2754 | 0.201 | 0.4484 | Yes |
| 58 | PSME3 | NA | 2797 | 0.199 | 0.4545 | Yes |
| 59 | IGKV1-17 | NA | 2814 | 0.199 | 0.4610 | Yes |
| 60 | IGHV4-39 | NA | 2816 | 0.199 | 0.4678 | Yes |
| 61 | IGKV1D-12 | NA | 2817 | 0.199 | 0.4747 | Yes |
| 62 | IGLC1 | NA | 2899 | 0.195 | 0.4799 | Yes |
| 63 | IGLV1-44 | NA | 3311 | 0.183 | 0.4787 | Yes |
| 64 | PSMA5 | NA | 3584 | 0.175 | 0.4797 | Yes |
| 65 | PSMD6 | NA | 3656 | 0.173 | 0.4844 | Yes |
| 66 | IGHV3-53 | NA | 3723 | 0.172 | 0.4891 | Yes |
| 67 | MS4A2 | NA | 3968 | 0.165 | 0.4903 | Yes |
| 68 | PSMD11 | NA | 4014 | 0.164 | 0.4951 | Yes |
| 69 | IGLV6-57 | NA | 4269 | 0.158 | 0.4958 | Yes |
| 70 | IGKV2-29 | NA | 4573 | 0.151 | 0.4955 | Yes |
| 71 | PSMA3 | NA | 4684 | 0.148 | 0.4986 | Yes |
| 72 | BTK | NA | 4928 | 0.143 | 0.4990 | Yes |
| 73 | LAT2 | NA | 5195 | 0.139 | 0.4989 | No |
| 74 | LCP2 | NA | 5521 | 0.132 | 0.4975 | No |
| 75 | PSMD5 | NA | 5798 | 0.126 | 0.4968 | No |
| 76 | FCER1A | NA | 6350 | 0.117 | 0.4907 | No |
| 77 | PSMA4 | NA | 6664 | 0.112 | 0.4888 | No |
| 78 | VAV3 | NA | 6667 | 0.112 | 0.4926 | No |
| 79 | VAV1 | NA | 7049 | 0.106 | 0.4892 | No |
| 80 | PAK1 | NA | 7092 | 0.105 | 0.4921 | No |
| 81 | NFATC2 | NA | 7189 | 0.104 | 0.4939 | No |
| 82 | IGLC7 | NA | 7419 | 0.100 | 0.4931 | No |
| 83 | PSMC4 | NA | 7603 | 0.097 | 0.4931 | No |
| 84 | MAPK1 | NA | 8583 | 0.083 | 0.4780 | No |
| 85 | CARD11 | NA | 8976 | 0.078 | 0.4735 | No |
| 86 | PSMD13 | NA | 8996 | 0.078 | 0.4759 | No |
| 87 | PSMB2 | NA | 9166 | 0.076 | 0.4754 | No |
| 88 | ITK | NA | 9525 | 0.072 | 0.4713 | No |
| 89 | PSMA8 | NA | 10060 | 0.065 | 0.4637 | No |
| 90 | CALM1 | NA | 10213 | 0.064 | 0.4631 | No |
| 91 | PLCG2 | NA | 10314 | 0.063 | 0.4634 | No |
| 92 | PSMD8 | NA | 11056 | 0.055 | 0.4517 | No |
| 93 | PSMB1 | NA | 11463 | 0.050 | 0.4460 | No |
| 94 | PSMB6 | NA | 11703 | 0.048 | 0.4432 | No |
| 95 | PSMF1 | NA | 12187 | 0.043 | 0.4358 | No |
| 96 | GRB2 | NA | 12411 | 0.041 | 0.4332 | No |
| 97 | PSMB5 | NA | 12955 | 0.037 | 0.4245 | No |
| 98 | NFATC3 | NA | 13319 | 0.034 | 0.4190 | No |
| 99 | SYK | NA | 14202 | 0.028 | 0.4037 | No |
| 100 | PSMD2 | NA | 14744 | 0.024 | 0.3946 | No |
| 101 | CHUK | NA | 14882 | 0.023 | 0.3929 | No |
| 102 | PAK2 | NA | 15011 | 0.022 | 0.3913 | No |
| 103 | PSMC3 | NA | 15166 | 0.021 | 0.3892 | No |
| 104 | PSMA2 | NA | 15393 | 0.020 | 0.3857 | No |
| 105 | PSMA1 | NA | 18778 | 0.003 | 0.3236 | No |
| 106 | PSMB9 | NA | 18922 | 0.002 | 0.3211 | No |
| 107 | PSMB7 | NA | 25656 | -0.005 | 0.1975 | No |
| 108 | BCL10 | NA | 26199 | -0.011 | 0.1879 | No |
| 109 | IKBKG | NA | 26607 | -0.015 | 0.1810 | No |
| 110 | FER | NA | 26807 | -0.016 | 0.1779 | No |
| 111 | NFATC1 | NA | 27642 | -0.024 | 0.1634 | No |
| 112 | PSME4 | NA | 27667 | -0.024 | 0.1638 | No |
| 113 | PPP3CB | NA | 27823 | -0.026 | 0.1618 | No |
| 114 | MAPK10 | NA | 27966 | -0.027 | 0.1601 | No |
| 115 | PSMB11 | NA | 28613 | -0.033 | 0.1494 | No |
| 116 | NR4A3 | NA | 29530 | -0.042 | 0.1340 | No |
| 117 | FOS | NA | 29699 | -0.044 | 0.1324 | No |
| 118 | PSMD4 | NA | 29970 | -0.046 | 0.1291 | No |
| 119 | PPP3CA | NA | 30263 | -0.049 | 0.1254 | No |
| 120 | NFKB1 | NA | 31293 | -0.060 | 0.1086 | No |
| 121 | GRAP2 | NA | 31627 | -0.063 | 0.1046 | No |
| 122 | PIK3CA | NA | 31724 | -0.064 | 0.1050 | No |
| 123 | SHC1 | NA | 32934 | -0.076 | 0.0854 | No |
| 124 | PSMB10 | NA | 33178 | -0.078 | 0.0837 | No |
| 125 | MAPK8 | NA | 33775 | -0.084 | 0.0756 | No |
| 126 | PSMB8 | NA | 34188 | -0.088 | 0.0711 | No |
| 127 | PSMA7 | NA | 34982 | -0.096 | 0.0598 | No |
| 128 | PSMC5 | NA | 35183 | -0.098 | 0.0595 | No |
| 129 | PPP3R1 | NA | 35383 | -0.100 | 0.0593 | No |
| 130 | PSMD3 | NA | 35676 | -0.103 | 0.0574 | No |
| 131 | JUN | NA | 35845 | -0.105 | 0.0580 | No |
| 132 | RAC1 | NA | 36138 | -0.107 | 0.0563 | No |
| 133 | PDPK1 | NA | 36179 | -0.108 | 0.0593 | No |
| 134 | CUL1 | NA | 36788 | -0.114 | 0.0520 | No |
| 135 | PSMD9 | NA | 37479 | -0.121 | 0.0435 | No |
| 136 | PSMB4 | NA | 38120 | -0.128 | 0.0361 | No |
| 137 | PSMD10 | NA | 38860 | -0.135 | 0.0272 | No |
| 138 | GAB2 | NA | 39696 | -0.145 | 0.0169 | No |
| 139 | MAP2K4 | NA | 39716 | -0.145 | 0.0215 | No |
| 140 | FBXW11 | NA | 40207 | -0.151 | 0.0177 | No |
| 141 | PRKCQ | NA | 40881 | -0.159 | 0.0108 | No |
| 142 | PSMB3 | NA | 40982 | -0.160 | 0.0145 | No |
| 143 | LYN | NA | 41372 | -0.165 | 0.0130 | No |
| 144 | MALT1 | NA | 42417 | -0.179 | -0.0000 | No |
| 145 | BTRC | NA | 43199 | -0.188 | -0.0079 | No |
| 146 | PSMC6 | NA | 43260 | -0.189 | -0.0025 | No |
| 147 | SOS1 | NA | 44034 | -0.200 | -0.0098 | No |
| 148 | MAP3K1 | NA | 44125 | -0.202 | -0.0045 | No |
| 149 | MAPK9 | NA | 44664 | -0.210 | -0.0072 | No |
| 150 | TAB2 | NA | 45196 | -0.217 | -0.0094 | No |
| 151 | PLCG1 | NA | 45668 | -0.225 | -0.0103 | No |
| 152 | TAB1 | NA | 46623 | -0.241 | -0.0196 | No |
| 153 | PIK3R2 | NA | 46657 | -0.241 | -0.0119 | No |
| 154 | SKP1 | NA | 46866 | -0.245 | -0.0073 | No |
| 155 | IKBKB | NA | 47515 | -0.256 | -0.0104 | No |
| 156 | TRAF6 | NA | 47561 | -0.257 | -0.0024 | No |
| 157 | TAB3 | NA | 47630 | -0.258 | 0.0053 | No |
| 158 | PSME1 | NA | 47870 | -0.263 | 0.0099 | No |
| 159 | MAPK3 | NA | 48613 | -0.278 | 0.0059 | No |
| 160 | PIK3CB | NA | 49238 | -0.292 | 0.0045 | No |
| 161 | PSME2 | NA | 49748 | -0.303 | 0.0055 | No |
| 162 | RELA | NA | 50168 | -0.313 | 0.0086 | No |
| 163 | MAP2K7 | NA | 50296 | -0.317 | 0.0172 | No |
| 164 | VAV2 | NA | 50383 | -0.319 | 0.0266 | No |
| 165 | TEC | NA | 50755 | -0.329 | 0.0311 | No |
| 166 | IGHE | NA | 51792 | -0.361 | 0.0245 | No |
| 167 | LAT | NA | 52157 | -0.374 | 0.0307 | No |
| 168 | MAP3K7 | NA | 52902 | -0.406 | 0.0310 | No |