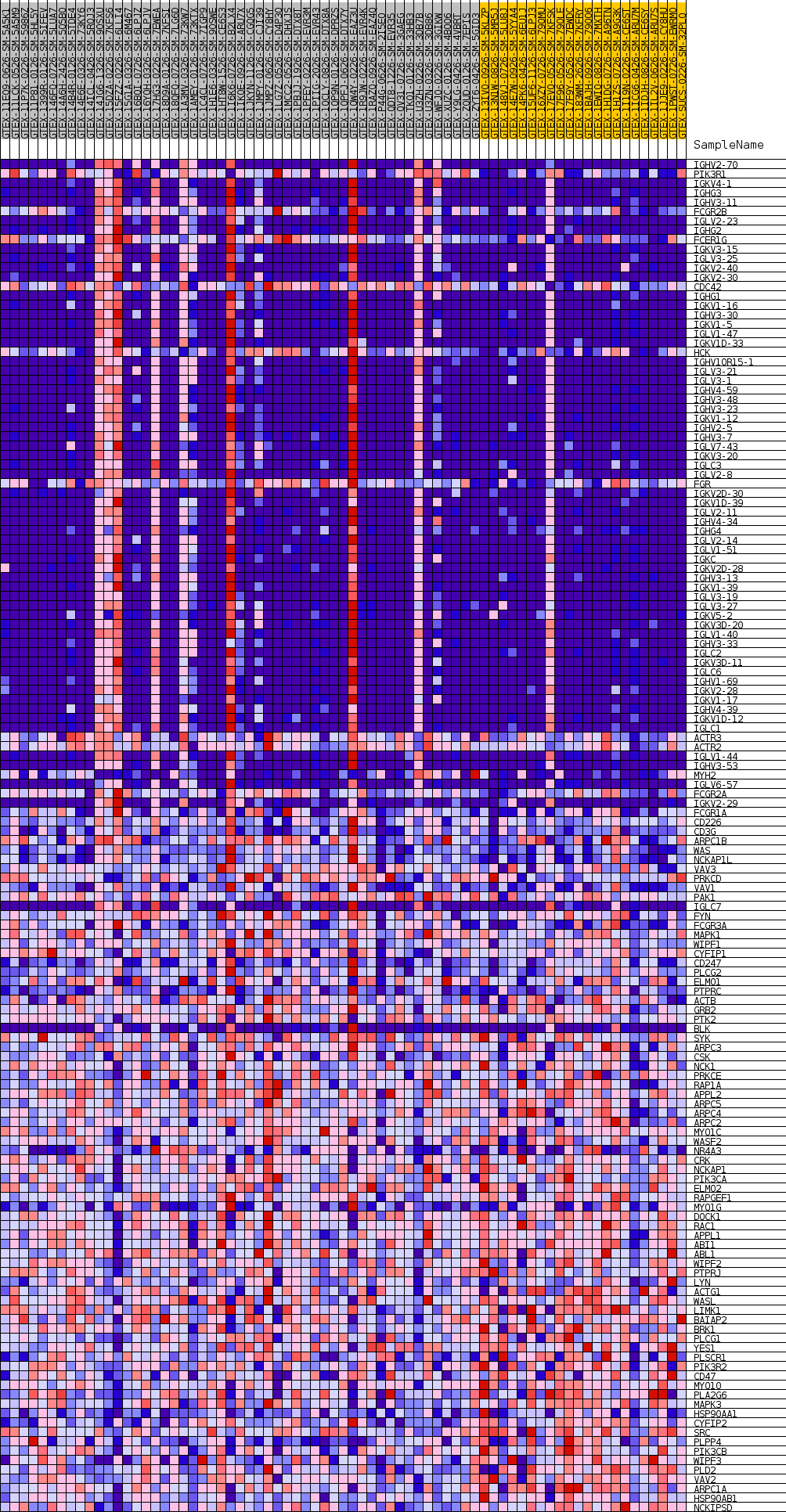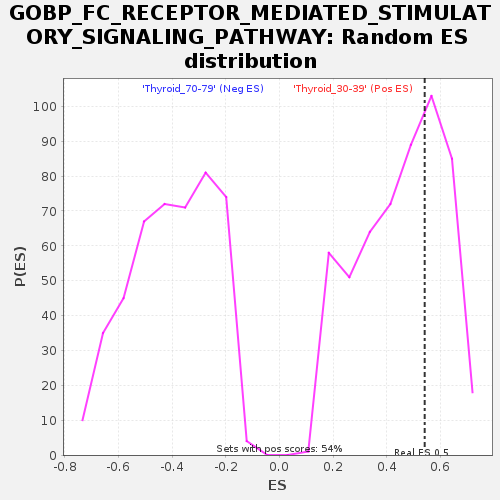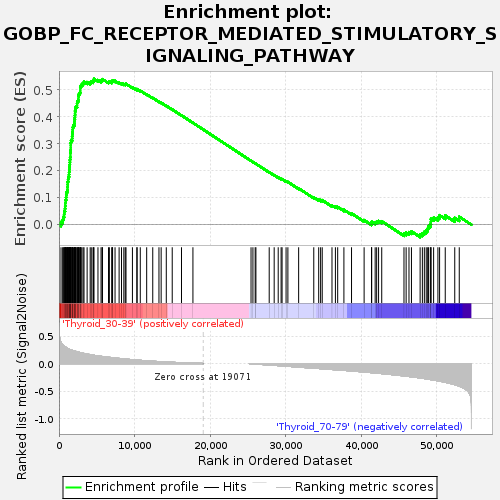
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Thyroid.Thyroid.cls #Thyroid_30-39_versus_Thyroid_70-79.Thyroid.cls #Thyroid_30-39_versus_Thyroid_70-79_repos |
| Phenotype | Thyroid.cls#Thyroid_30-39_versus_Thyroid_70-79_repos |
| Upregulated in class | Thyroid_30-39 |
| GeneSet | GOBP_FC_RECEPTOR_MEDIATED_STIMULATORY_SIGNALING_PATHWAY |
| Enrichment Score (ES) | 0.5411125 |
| Normalized Enrichment Score (NES) | 1.195836 |
| Nominal p-value | 0.34750462 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | IGHV2-70 | NA | 266 | 0.384 | 0.0094 | Yes |
| 2 | PIK3R1 | NA | 488 | 0.341 | 0.0180 | Yes |
| 3 | IGKV4-1 | NA | 570 | 0.329 | 0.0288 | Yes |
| 4 | IGHG3 | NA | 685 | 0.317 | 0.0384 | Yes |
| 5 | IGHV3-11 | NA | 702 | 0.315 | 0.0498 | Yes |
| 6 | FCGR2B | NA | 799 | 0.305 | 0.0594 | Yes |
| 7 | IGLV2-23 | NA | 825 | 0.303 | 0.0702 | Yes |
| 8 | IGHG2 | NA | 846 | 0.301 | 0.0810 | Yes |
| 9 | FCER1G | NA | 868 | 0.298 | 0.0917 | Yes |
| 10 | IGKV3-15 | NA | 938 | 0.291 | 0.1013 | Yes |
| 11 | IGLV3-25 | NA | 962 | 0.289 | 0.1116 | Yes |
| 12 | IGKV2-40 | NA | 1015 | 0.284 | 0.1212 | Yes |
| 13 | IGKV2-30 | NA | 1114 | 0.276 | 0.1296 | Yes |
| 14 | CDC42 | NA | 1134 | 0.274 | 0.1395 | Yes |
| 15 | IGHG1 | NA | 1146 | 0.273 | 0.1494 | Yes |
| 16 | IGKV1-16 | NA | 1158 | 0.272 | 0.1593 | Yes |
| 17 | IGHV3-30 | NA | 1213 | 0.269 | 0.1683 | Yes |
| 18 | IGKV1-5 | NA | 1241 | 0.267 | 0.1778 | Yes |
| 19 | IGLV1-47 | NA | 1327 | 0.261 | 0.1859 | Yes |
| 20 | IGKV1D-33 | NA | 1341 | 0.260 | 0.1953 | Yes |
| 21 | HCK | NA | 1361 | 0.259 | 0.2046 | Yes |
| 22 | IGHV1OR15-1 | NA | 1378 | 0.258 | 0.2139 | Yes |
| 23 | IGLV3-21 | NA | 1420 | 0.255 | 0.2226 | Yes |
| 24 | IGLV3-1 | NA | 1435 | 0.255 | 0.2318 | Yes |
| 25 | IGHV4-59 | NA | 1451 | 0.254 | 0.2410 | Yes |
| 26 | IGHV3-48 | NA | 1478 | 0.252 | 0.2499 | Yes |
| 27 | IGHV3-23 | NA | 1510 | 0.251 | 0.2587 | Yes |
| 28 | IGKV1-12 | NA | 1511 | 0.251 | 0.2680 | Yes |
| 29 | IGHV2-5 | NA | 1536 | 0.249 | 0.2768 | Yes |
| 30 | IGHV3-7 | NA | 1550 | 0.248 | 0.2858 | Yes |
| 31 | IGLV7-43 | NA | 1552 | 0.248 | 0.2950 | Yes |
| 32 | IGKV3-20 | NA | 1555 | 0.248 | 0.3042 | Yes |
| 33 | IGLC3 | NA | 1602 | 0.246 | 0.3124 | Yes |
| 34 | IGLV2-8 | NA | 1729 | 0.239 | 0.3190 | Yes |
| 35 | FGR | NA | 1741 | 0.239 | 0.3277 | Yes |
| 36 | IGKV2D-30 | NA | 1768 | 0.238 | 0.3360 | Yes |
| 37 | IGKV1D-39 | NA | 1794 | 0.236 | 0.3444 | Yes |
| 38 | IGLV2-11 | NA | 1811 | 0.236 | 0.3528 | Yes |
| 39 | IGHV4-34 | NA | 1818 | 0.235 | 0.3615 | Yes |
| 40 | IGHG4 | NA | 1903 | 0.232 | 0.3685 | Yes |
| 41 | IGLV2-14 | NA | 2033 | 0.227 | 0.3746 | Yes |
| 42 | IGLV1-51 | NA | 2044 | 0.227 | 0.3828 | Yes |
| 43 | IGKC | NA | 2057 | 0.226 | 0.3910 | Yes |
| 44 | IGKV2D-28 | NA | 2099 | 0.225 | 0.3986 | Yes |
| 45 | IGHV3-13 | NA | 2110 | 0.224 | 0.4068 | Yes |
| 46 | IGKV1-39 | NA | 2126 | 0.224 | 0.4148 | Yes |
| 47 | IGLV3-19 | NA | 2132 | 0.224 | 0.4230 | Yes |
| 48 | IGLV3-27 | NA | 2209 | 0.221 | 0.4299 | Yes |
| 49 | IGKV5-2 | NA | 2231 | 0.220 | 0.4376 | Yes |
| 50 | IGKV3D-20 | NA | 2398 | 0.213 | 0.4425 | Yes |
| 51 | IGLV1-40 | NA | 2400 | 0.213 | 0.4504 | Yes |
| 52 | IGHV3-33 | NA | 2419 | 0.212 | 0.4579 | Yes |
| 53 | IGLC2 | NA | 2547 | 0.208 | 0.4633 | Yes |
| 54 | IGKV3D-11 | NA | 2573 | 0.207 | 0.4706 | Yes |
| 55 | IGLC6 | NA | 2574 | 0.207 | 0.4783 | Yes |
| 56 | IGHV1-69 | NA | 2627 | 0.205 | 0.4849 | Yes |
| 57 | IGKV2-28 | NA | 2754 | 0.201 | 0.4901 | Yes |
| 58 | IGKV1-17 | NA | 2814 | 0.199 | 0.4963 | Yes |
| 59 | IGHV4-39 | NA | 2816 | 0.199 | 0.5037 | Yes |
| 60 | IGKV1D-12 | NA | 2817 | 0.199 | 0.5111 | Yes |
| 61 | IGLC1 | NA | 2899 | 0.195 | 0.5169 | Yes |
| 62 | ACTR3 | NA | 3019 | 0.191 | 0.5218 | Yes |
| 63 | ACTR2 | NA | 3191 | 0.186 | 0.5256 | Yes |
| 64 | IGLV1-44 | NA | 3311 | 0.183 | 0.5302 | Yes |
| 65 | IGHV3-53 | NA | 3723 | 0.172 | 0.5290 | Yes |
| 66 | MYH2 | NA | 4127 | 0.161 | 0.5276 | Yes |
| 67 | IGLV6-57 | NA | 4269 | 0.158 | 0.5309 | Yes |
| 68 | FCGR2A | NA | 4504 | 0.152 | 0.5322 | Yes |
| 69 | IGKV2-29 | NA | 4573 | 0.151 | 0.5366 | Yes |
| 70 | FCGR1A | NA | 4630 | 0.149 | 0.5411 | Yes |
| 71 | CD226 | NA | 5158 | 0.139 | 0.5366 | No |
| 72 | CD3G | NA | 5566 | 0.131 | 0.5340 | No |
| 73 | ARPC1B | NA | 5709 | 0.128 | 0.5362 | No |
| 74 | WAS | NA | 5760 | 0.127 | 0.5400 | No |
| 75 | NCKAP1L | NA | 6555 | 0.114 | 0.5296 | No |
| 76 | VAV3 | NA | 6667 | 0.112 | 0.5317 | No |
| 77 | PRKCD | NA | 6985 | 0.107 | 0.5299 | No |
| 78 | VAV1 | NA | 7049 | 0.106 | 0.5327 | No |
| 79 | PAK1 | NA | 7092 | 0.105 | 0.5358 | No |
| 80 | IGLC7 | NA | 7419 | 0.100 | 0.5336 | No |
| 81 | FYN | NA | 7957 | 0.092 | 0.5271 | No |
| 82 | FCGR3A | NA | 8275 | 0.088 | 0.5246 | No |
| 83 | MAPK1 | NA | 8583 | 0.083 | 0.5220 | No |
| 84 | WIPF1 | NA | 8768 | 0.081 | 0.5217 | No |
| 85 | CYFIP1 | NA | 8869 | 0.080 | 0.5228 | No |
| 86 | CD247 | NA | 9720 | 0.069 | 0.5098 | No |
| 87 | PLCG2 | NA | 10314 | 0.063 | 0.5012 | No |
| 88 | ELMO1 | NA | 10332 | 0.062 | 0.5032 | No |
| 89 | PTPRC | NA | 10760 | 0.058 | 0.4975 | No |
| 90 | ACTB | NA | 11607 | 0.049 | 0.4838 | No |
| 91 | GRB2 | NA | 12411 | 0.041 | 0.4706 | No |
| 92 | PTK2 | NA | 13232 | 0.035 | 0.4568 | No |
| 93 | BLK | NA | 13523 | 0.033 | 0.4527 | No |
| 94 | SYK | NA | 14202 | 0.028 | 0.4413 | No |
| 95 | ARPC3 | NA | 14988 | 0.022 | 0.4277 | No |
| 96 | CSK | NA | 16216 | 0.015 | 0.4057 | No |
| 97 | NCK1 | NA | 17729 | 0.007 | 0.3782 | No |
| 98 | PRKCE | NA | 25421 | -0.003 | 0.2370 | No |
| 99 | RAP1A | NA | 25671 | -0.005 | 0.2327 | No |
| 100 | APPL2 | NA | 25968 | -0.009 | 0.2275 | No |
| 101 | ARPC5 | NA | 26067 | -0.010 | 0.2261 | No |
| 102 | ARPC4 | NA | 27827 | -0.026 | 0.1947 | No |
| 103 | ARPC2 | NA | 28485 | -0.032 | 0.1839 | No |
| 104 | MYO1C | NA | 29019 | -0.037 | 0.1754 | No |
| 105 | WASF2 | NA | 29402 | -0.041 | 0.1699 | No |
| 106 | NR4A3 | NA | 29530 | -0.042 | 0.1692 | No |
| 107 | CRK | NA | 30059 | -0.047 | 0.1612 | No |
| 108 | NCKAP1 | NA | 30300 | -0.050 | 0.1587 | No |
| 109 | PIK3CA | NA | 31724 | -0.064 | 0.1349 | No |
| 110 | ELMO2 | NA | 33726 | -0.083 | 0.1013 | No |
| 111 | RAPGEF1 | NA | 34378 | -0.090 | 0.0926 | No |
| 112 | MYO1G | NA | 34633 | -0.093 | 0.0914 | No |
| 113 | DOCK1 | NA | 34846 | -0.095 | 0.0910 | No |
| 114 | RAC1 | NA | 36138 | -0.107 | 0.0713 | No |
| 115 | APPL1 | NA | 36590 | -0.112 | 0.0672 | No |
| 116 | ABI1 | NA | 36892 | -0.115 | 0.0659 | No |
| 117 | ABL1 | NA | 37715 | -0.123 | 0.0554 | No |
| 118 | WIPF2 | NA | 38725 | -0.134 | 0.0418 | No |
| 119 | PTPRJ | NA | 40391 | -0.153 | 0.0170 | No |
| 120 | LYN | NA | 41372 | -0.165 | 0.0051 | No |
| 121 | ACTG1 | NA | 41393 | -0.165 | 0.0109 | No |
| 122 | WASL | NA | 41853 | -0.171 | 0.0088 | No |
| 123 | LIMK1 | NA | 42051 | -0.174 | 0.0116 | No |
| 124 | BAIAP2 | NA | 42306 | -0.177 | 0.0136 | No |
| 125 | BRK1 | NA | 42723 | -0.182 | 0.0127 | No |
| 126 | PLCG1 | NA | 45668 | -0.225 | -0.0330 | No |
| 127 | YES1 | NA | 45953 | -0.230 | -0.0297 | No |
| 128 | PLSCR1 | NA | 46330 | -0.235 | -0.0279 | No |
| 129 | PIK3R2 | NA | 46657 | -0.241 | -0.0249 | No |
| 130 | CD47 | NA | 47805 | -0.262 | -0.0363 | No |
| 131 | MYO10 | NA | 48102 | -0.267 | -0.0318 | No |
| 132 | PLA2G6 | NA | 48363 | -0.273 | -0.0264 | No |
| 133 | MAPK3 | NA | 48613 | -0.278 | -0.0206 | No |
| 134 | HSP90AA1 | NA | 48810 | -0.282 | -0.0138 | No |
| 135 | CYFIP2 | NA | 48905 | -0.285 | -0.0049 | No |
| 136 | SRC | NA | 49154 | -0.290 | 0.0013 | No |
| 137 | PLPP4 | NA | 49229 | -0.292 | 0.0108 | No |
| 138 | PIK3CB | NA | 49238 | -0.292 | 0.0215 | No |
| 139 | WIPF3 | NA | 49592 | -0.300 | 0.0261 | No |
| 140 | PLD2 | NA | 50160 | -0.313 | 0.0274 | No |
| 141 | VAV2 | NA | 50383 | -0.319 | 0.0351 | No |
| 142 | ARPC1A | NA | 51131 | -0.340 | 0.0341 | No |
| 143 | HSP90AB1 | NA | 52389 | -0.382 | 0.0252 | No |
| 144 | NCKIPSD | NA | 52986 | -0.410 | 0.0295 | No |