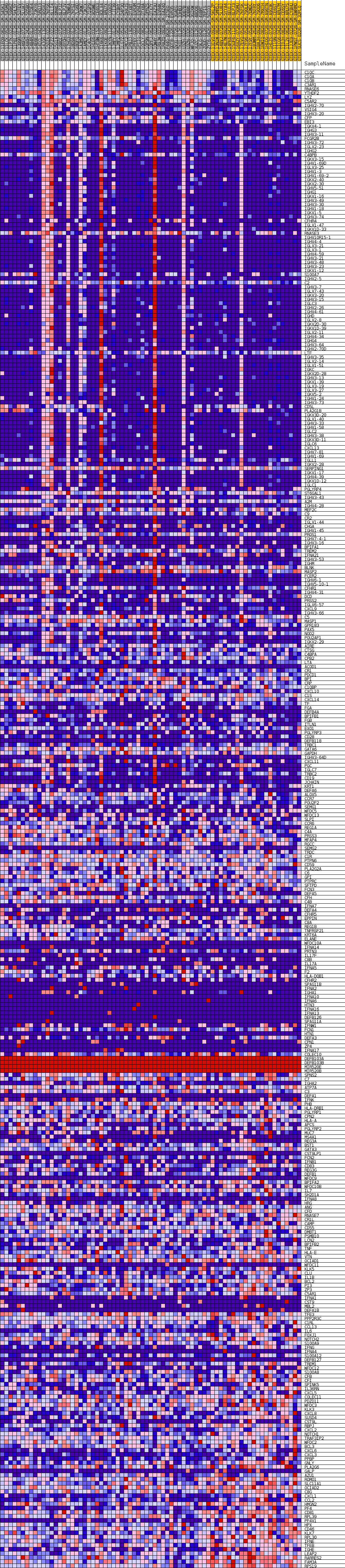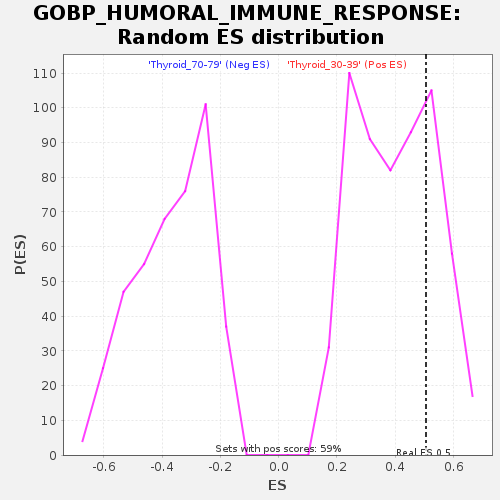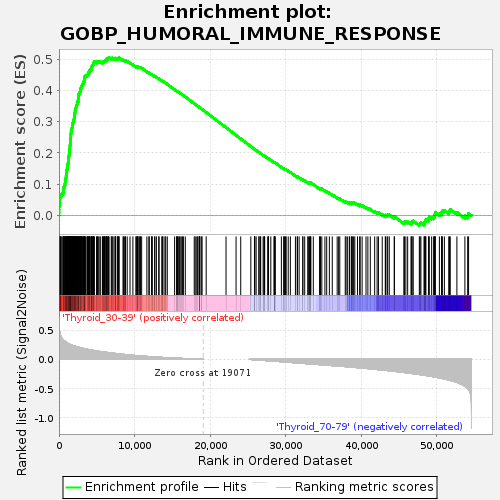
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Thyroid.Thyroid.cls #Thyroid_30-39_versus_Thyroid_70-79.Thyroid.cls #Thyroid_30-39_versus_Thyroid_70-79_repos |
| Phenotype | Thyroid.cls#Thyroid_30-39_versus_Thyroid_70-79_repos |
| Upregulated in class | Thyroid_30-39 |
| GeneSet | GOBP_HUMORAL_IMMUNE_RESPONSE |
| Enrichment Score (ES) | 0.50546587 |
| Normalized Enrichment Score (NES) | 1.2641248 |
| Nominal p-value | 0.2640545 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | C1QC | NA | 22 | 0.551 | 0.0089 | Yes |
| 2 | C1QA | NA | 28 | 0.541 | 0.0179 | Yes |
| 3 | C1QB | NA | 33 | 0.534 | 0.0269 | Yes |
| 4 | C3AR1 | NA | 46 | 0.512 | 0.0353 | Yes |
| 5 | RNASE6 | NA | 92 | 0.466 | 0.0424 | Yes |
| 6 | YTHDF2 | NA | 156 | 0.424 | 0.0483 | Yes |
| 7 | LYZ | NA | 157 | 0.424 | 0.0555 | Yes |
| 8 | C5AR2 | NA | 167 | 0.419 | 0.0624 | Yes |
| 9 | IGHV2-70 | NA | 266 | 0.384 | 0.0671 | Yes |
| 10 | VSIG4 | NA | 383 | 0.359 | 0.0710 | Yes |
| 11 | IGHV3-20 | NA | 541 | 0.333 | 0.0738 | Yes |
| 12 | CFP | NA | 562 | 0.330 | 0.0790 | Yes |
| 13 | EBI3 | NA | 568 | 0.330 | 0.0844 | Yes |
| 14 | IGKV4-1 | NA | 570 | 0.329 | 0.0900 | Yes |
| 15 | IGHG3 | NA | 685 | 0.317 | 0.0932 | Yes |
| 16 | IGHV3-11 | NA | 702 | 0.315 | 0.0982 | Yes |
| 17 | FCGR2B | NA | 799 | 0.305 | 0.1016 | Yes |
| 18 | IGHV3-72 | NA | 821 | 0.303 | 0.1063 | Yes |
| 19 | IGLV2-23 | NA | 825 | 0.303 | 0.1114 | Yes |
| 20 | IGHG2 | NA | 846 | 0.301 | 0.1161 | Yes |
| 21 | C4BPB | NA | 906 | 0.294 | 0.1200 | Yes |
| 22 | IGKV3-15 | NA | 938 | 0.291 | 0.1243 | Yes |
| 23 | IGHV1-69D | NA | 947 | 0.290 | 0.1291 | Yes |
| 24 | IGLV3-25 | NA | 962 | 0.289 | 0.1337 | Yes |
| 25 | IGHV1-3 | NA | 1000 | 0.286 | 0.1378 | Yes |
| 26 | IGHV1-69-2 | NA | 1014 | 0.284 | 0.1424 | Yes |
| 27 | IGKV2-40 | NA | 1015 | 0.284 | 0.1472 | Yes |
| 28 | IGKV2-30 | NA | 1114 | 0.276 | 0.1500 | Yes |
| 29 | IGHV5-51 | NA | 1135 | 0.274 | 0.1543 | Yes |
| 30 | IGHG1 | NA | 1146 | 0.273 | 0.1587 | Yes |
| 31 | IGKV1-16 | NA | 1158 | 0.272 | 0.1631 | Yes |
| 32 | IGHV3-49 | NA | 1199 | 0.270 | 0.1670 | Yes |
| 33 | IGHV3-30 | NA | 1213 | 0.269 | 0.1713 | Yes |
| 34 | IGHV1-18 | NA | 1236 | 0.267 | 0.1754 | Yes |
| 35 | IGKV1-5 | NA | 1241 | 0.267 | 0.1798 | Yes |
| 36 | IGHV3-74 | NA | 1243 | 0.267 | 0.1843 | Yes |
| 37 | CFHR4 | NA | 1268 | 0.265 | 0.1883 | Yes |
| 38 | IGLV1-47 | NA | 1327 | 0.261 | 0.1916 | Yes |
| 39 | IGKV1D-33 | NA | 1341 | 0.260 | 0.1958 | Yes |
| 40 | RNASE3 | NA | 1348 | 0.260 | 0.2001 | Yes |
| 41 | IGHV1OR15-1 | NA | 1378 | 0.258 | 0.2039 | Yes |
| 42 | IGHV4-4 | NA | 1379 | 0.258 | 0.2083 | Yes |
| 43 | IGLV3-21 | NA | 1420 | 0.255 | 0.2118 | Yes |
| 44 | IGLV3-1 | NA | 1435 | 0.255 | 0.2159 | Yes |
| 45 | IGHV4-59 | NA | 1451 | 0.254 | 0.2199 | Yes |
| 46 | IGHV3-21 | NA | 1460 | 0.253 | 0.2240 | Yes |
| 47 | IGHV3-48 | NA | 1478 | 0.252 | 0.2279 | Yes |
| 48 | IGHV3-23 | NA | 1510 | 0.251 | 0.2316 | Yes |
| 49 | IGKV1-12 | NA | 1511 | 0.251 | 0.2358 | Yes |
| 50 | S100A7 | NA | 1533 | 0.249 | 0.2397 | Yes |
| 51 | IGHV2-5 | NA | 1536 | 0.249 | 0.2438 | Yes |
| 52 | C2 | NA | 1543 | 0.249 | 0.2479 | Yes |
| 53 | IGHV3-7 | NA | 1550 | 0.248 | 0.2520 | Yes |
| 54 | IGLV7-43 | NA | 1552 | 0.248 | 0.2562 | Yes |
| 55 | IGKV3-20 | NA | 1555 | 0.248 | 0.2603 | Yes |
| 56 | IGHV3-15 | NA | 1586 | 0.246 | 0.2639 | Yes |
| 57 | IGLC3 | NA | 1602 | 0.246 | 0.2678 | Yes |
| 58 | IGHV2-26 | NA | 1612 | 0.245 | 0.2718 | Yes |
| 59 | IGHV4-61 | NA | 1690 | 0.242 | 0.2744 | Yes |
| 60 | IGHD | NA | 1697 | 0.241 | 0.2784 | Yes |
| 61 | IGLV2-8 | NA | 1729 | 0.239 | 0.2819 | Yes |
| 62 | IGKV2D-30 | NA | 1768 | 0.238 | 0.2852 | Yes |
| 63 | IGKV1D-39 | NA | 1794 | 0.236 | 0.2887 | Yes |
| 64 | IGLV2-11 | NA | 1811 | 0.236 | 0.2924 | Yes |
| 65 | IGHV4-34 | NA | 1818 | 0.235 | 0.2962 | Yes |
| 66 | IGHG4 | NA | 1903 | 0.232 | 0.2986 | Yes |
| 67 | IGHV3-64 | NA | 1932 | 0.231 | 0.3020 | Yes |
| 68 | IGHV2-70D | NA | 1979 | 0.229 | 0.3050 | Yes |
| 69 | LTF | NA | 1983 | 0.229 | 0.3088 | Yes |
| 70 | IGHV3-35 | NA | 2011 | 0.228 | 0.3122 | Yes |
| 71 | IGLV2-14 | NA | 2033 | 0.227 | 0.3156 | Yes |
| 72 | IGLV1-51 | NA | 2044 | 0.227 | 0.3192 | Yes |
| 73 | IGKC | NA | 2057 | 0.226 | 0.3228 | Yes |
| 74 | IGKV2D-28 | NA | 2099 | 0.225 | 0.3259 | Yes |
| 75 | IGHV3-13 | NA | 2110 | 0.224 | 0.3295 | Yes |
| 76 | IGKV1-39 | NA | 2126 | 0.224 | 0.3330 | Yes |
| 77 | IGLV3-19 | NA | 2132 | 0.224 | 0.3367 | Yes |
| 78 | IGLV3-27 | NA | 2209 | 0.221 | 0.3390 | Yes |
| 79 | IGKV5-2 | NA | 2231 | 0.220 | 0.3423 | Yes |
| 80 | IGHV1-24 | NA | 2244 | 0.219 | 0.3458 | Yes |
| 81 | IGHV3-73 | NA | 2283 | 0.217 | 0.3488 | Yes |
| 82 | CD5L | NA | 2335 | 0.215 | 0.3514 | Yes |
| 83 | PLA2G1B | NA | 2366 | 0.214 | 0.3545 | Yes |
| 84 | IGKV3D-20 | NA | 2398 | 0.213 | 0.3575 | Yes |
| 85 | IGLV1-40 | NA | 2400 | 0.213 | 0.3611 | Yes |
| 86 | IGHV3-33 | NA | 2419 | 0.212 | 0.3644 | Yes |
| 87 | IGHV1-58 | NA | 2539 | 0.208 | 0.3657 | Yes |
| 88 | IGLC2 | NA | 2547 | 0.208 | 0.3691 | Yes |
| 89 | IGHV3-38 | NA | 2548 | 0.208 | 0.3726 | Yes |
| 90 | IGKV3D-11 | NA | 2573 | 0.207 | 0.3756 | Yes |
| 91 | IGLC6 | NA | 2574 | 0.207 | 0.3791 | Yes |
| 92 | CXCL13 | NA | 2612 | 0.205 | 0.3819 | Yes |
| 93 | IGHV7-81 | NA | 2622 | 0.205 | 0.3852 | Yes |
| 94 | IGHV1-69 | NA | 2627 | 0.205 | 0.3886 | Yes |
| 95 | IGLL1 | NA | 2697 | 0.202 | 0.3907 | Yes |
| 96 | IGKV2-28 | NA | 2754 | 0.201 | 0.3931 | Yes |
| 97 | SERPING1 | NA | 2784 | 0.199 | 0.3959 | Yes |
| 98 | IGKV1-17 | NA | 2814 | 0.199 | 0.3987 | Yes |
| 99 | IGHV4-39 | NA | 2816 | 0.199 | 0.4021 | Yes |
| 100 | IGKV1D-12 | NA | 2817 | 0.199 | 0.4054 | Yes |
| 101 | IGLC1 | NA | 2899 | 0.195 | 0.4072 | Yes |
| 102 | PGLYRP4 | NA | 2962 | 0.193 | 0.4093 | Yes |
| 103 | ST6GAL1 | NA | 2979 | 0.193 | 0.4123 | Yes |
| 104 | IGHV3-43 | NA | 3029 | 0.191 | 0.4146 | Yes |
| 105 | A2M | NA | 3063 | 0.190 | 0.4172 | Yes |
| 106 | IGHV4-28 | NA | 3075 | 0.190 | 0.4202 | Yes |
| 107 | MEF2C | NA | 3185 | 0.186 | 0.4214 | Yes |
| 108 | C9 | NA | 3242 | 0.185 | 0.4234 | Yes |
| 109 | CR2 | NA | 3270 | 0.184 | 0.4261 | Yes |
| 110 | IGLV1-44 | NA | 3311 | 0.183 | 0.4284 | Yes |
| 111 | CHGA | NA | 3323 | 0.183 | 0.4313 | Yes |
| 112 | IGHV1-45 | NA | 3364 | 0.182 | 0.4336 | Yes |
| 113 | PROS1 | NA | 3365 | 0.182 | 0.4367 | Yes |
| 114 | IGHV7-4-1 | NA | 3393 | 0.181 | 0.4392 | Yes |
| 115 | IGHV3-16 | NA | 3394 | 0.181 | 0.4423 | Yes |
| 116 | BPIFA1 | NA | 3403 | 0.181 | 0.4452 | Yes |
| 117 | TREM2 | NA | 3542 | 0.176 | 0.4456 | Yes |
| 118 | IFNA21 | NA | 3545 | 0.176 | 0.4486 | Yes |
| 119 | IGHV3-53 | NA | 3723 | 0.172 | 0.4482 | Yes |
| 120 | IGHM | NA | 3785 | 0.170 | 0.4499 | Yes |
| 121 | BLNK | NA | 3853 | 0.168 | 0.4515 | Yes |
| 122 | MASP2 | NA | 3857 | 0.168 | 0.4543 | Yes |
| 123 | FCER2 | NA | 3869 | 0.168 | 0.4570 | Yes |
| 124 | IGHV6-1 | NA | 3991 | 0.165 | 0.4575 | Yes |
| 125 | IGHV5-10-1 | NA | 4000 | 0.164 | 0.4601 | Yes |
| 126 | CFHR1 | NA | 4045 | 0.163 | 0.4621 | Yes |
| 127 | IGHV4-31 | NA | 4069 | 0.163 | 0.4644 | Yes |
| 128 | DCD | NA | 4115 | 0.161 | 0.4663 | Yes |
| 129 | PRSS2 | NA | 4261 | 0.158 | 0.4663 | Yes |
| 130 | IGLV6-57 | NA | 4269 | 0.158 | 0.4688 | Yes |
| 131 | CXCL9 | NA | 4288 | 0.157 | 0.4711 | Yes |
| 132 | IGHV3-66 | NA | 4327 | 0.156 | 0.4731 | Yes |
| 133 | C5 | NA | 4330 | 0.156 | 0.4757 | Yes |
| 134 | MASP1 | NA | 4355 | 0.156 | 0.4779 | Yes |
| 135 | GPR183 | NA | 4391 | 0.155 | 0.4798 | Yes |
| 136 | PAX5 | NA | 4488 | 0.153 | 0.4806 | Yes |
| 137 | NOD2 | NA | 4498 | 0.152 | 0.4830 | Yes |
| 138 | POU2AF1 | NA | 4559 | 0.151 | 0.4845 | Yes |
| 139 | IGKV2-29 | NA | 4573 | 0.151 | 0.4868 | Yes |
| 140 | AIRE | NA | 4603 | 0.150 | 0.4888 | Yes |
| 141 | CTSG | NA | 4631 | 0.149 | 0.4908 | Yes |
| 142 | C4BPA | NA | 4672 | 0.149 | 0.4926 | Yes |
| 143 | CPB2 | NA | 4984 | 0.143 | 0.4892 | Yes |
| 144 | LTA | NA | 5001 | 0.142 | 0.4914 | Yes |
| 145 | ACOD1 | NA | 5098 | 0.140 | 0.4920 | Yes |
| 146 | CR1 | NA | 5194 | 0.139 | 0.4925 | Yes |
| 147 | PDCD1 | NA | 5319 | 0.136 | 0.4925 | Yes |
| 148 | BPI | NA | 5515 | 0.132 | 0.4912 | Yes |
| 149 | CRP | NA | 5763 | 0.127 | 0.4888 | Yes |
| 150 | C1QBP | NA | 5767 | 0.127 | 0.4909 | Yes |
| 151 | CXCL10 | NA | 5876 | 0.125 | 0.4910 | Yes |
| 152 | C1S | NA | 5931 | 0.124 | 0.4921 | Yes |
| 153 | CXCL14 | NA | 5995 | 0.123 | 0.4930 | Yes |
| 154 | TF | NA | 6027 | 0.123 | 0.4945 | Yes |
| 155 | FGA | NA | 6108 | 0.121 | 0.4951 | Yes |
| 156 | DEFB4A | NA | 6209 | 0.120 | 0.4953 | Yes |
| 157 | BPIFB1 | NA | 6274 | 0.118 | 0.4961 | Yes |
| 158 | FGB | NA | 6275 | 0.118 | 0.4981 | Yes |
| 159 | ITLN1 | NA | 6286 | 0.118 | 0.4999 | Yes |
| 160 | EXO1 | NA | 6298 | 0.118 | 0.5017 | Yes |
| 161 | PGLYRP3 | NA | 6455 | 0.116 | 0.5008 | Yes |
| 162 | CD28 | NA | 6477 | 0.115 | 0.5023 | Yes |
| 163 | DEFB118 | NA | 6515 | 0.114 | 0.5036 | Yes |
| 164 | TRBC1 | NA | 6565 | 0.114 | 0.5046 | Yes |
| 165 | GATA6 | NA | 6621 | 0.113 | 0.5055 | Yes |
| 166 | GAPDH | NA | 6913 | 0.108 | 0.5019 | No |
| 167 | IGHV3-64D | NA | 7005 | 0.107 | 0.5020 | No |
| 168 | CXCL11 | NA | 7029 | 0.106 | 0.5034 | No |
| 169 | PGC | NA | 7162 | 0.104 | 0.5027 | No |
| 170 | IGLC7 | NA | 7419 | 0.100 | 0.4997 | No |
| 171 | TRBC2 | NA | 7436 | 0.100 | 0.5011 | No |
| 172 | CD19 | NA | 7476 | 0.099 | 0.5021 | No |
| 173 | JCHAIN | NA | 7504 | 0.099 | 0.5032 | No |
| 174 | KRT1 | NA | 7728 | 0.096 | 0.5007 | No |
| 175 | DEFA6 | NA | 7799 | 0.095 | 0.5010 | No |
| 176 | ALOX5 | NA | 7833 | 0.094 | 0.5020 | No |
| 177 | CCR7 | NA | 7857 | 0.094 | 0.5032 | No |
| 178 | POU2F2 | NA | 7907 | 0.093 | 0.5038 | No |
| 179 | SEMG1 | NA | 7962 | 0.092 | 0.5044 | No |
| 180 | WFDC5 | NA | 8480 | 0.085 | 0.4963 | No |
| 181 | WFDC13 | NA | 8526 | 0.084 | 0.4969 | No |
| 182 | SLPI | NA | 8698 | 0.082 | 0.4951 | No |
| 183 | CCR6 | NA | 8773 | 0.081 | 0.4951 | No |
| 184 | REG1A | NA | 8811 | 0.080 | 0.4958 | No |
| 185 | C4A | NA | 9029 | 0.078 | 0.4931 | No |
| 186 | PRSS3 | NA | 9399 | 0.073 | 0.4875 | No |
| 187 | MFAP4 | NA | 9782 | 0.069 | 0.4817 | No |
| 188 | RGCC | NA | 10193 | 0.064 | 0.4752 | No |
| 189 | SEMG2 | NA | 10218 | 0.064 | 0.4758 | No |
| 190 | TRDC | NA | 10333 | 0.062 | 0.4747 | No |
| 191 | C1R | NA | 10343 | 0.062 | 0.4756 | No |
| 192 | PTPN6 | NA | 10393 | 0.062 | 0.4758 | No |
| 193 | CD59 | NA | 10439 | 0.061 | 0.4760 | No |
| 194 | PLA2G2A | NA | 10572 | 0.060 | 0.4745 | No |
| 195 | C6 | NA | 10692 | 0.058 | 0.4733 | No |
| 196 | GPI | NA | 10758 | 0.058 | 0.4731 | No |
| 197 | PTPRC | NA | 10760 | 0.058 | 0.4741 | No |
| 198 | SFTPD | NA | 10942 | 0.056 | 0.4717 | No |
| 199 | FCN3 | NA | 11632 | 0.049 | 0.4598 | No |
| 200 | DEFA5 | NA | 11898 | 0.046 | 0.4557 | No |
| 201 | CFH | NA | 11950 | 0.046 | 0.4555 | No |
| 202 | C4B | NA | 12255 | 0.043 | 0.4506 | No |
| 203 | IFNA7 | NA | 12319 | 0.042 | 0.4502 | No |
| 204 | DEFA4 | NA | 12325 | 0.042 | 0.4508 | No |
| 205 | CFHR5 | NA | 12663 | 0.039 | 0.4453 | No |
| 206 | EPPIN | NA | 12690 | 0.039 | 0.4454 | No |
| 207 | C8A | NA | 12895 | 0.038 | 0.4423 | No |
| 208 | REG1B | NA | 13207 | 0.035 | 0.4372 | No |
| 209 | TNFRSF21 | NA | 13245 | 0.035 | 0.4371 | No |
| 210 | KRT6A | NA | 13606 | 0.032 | 0.4310 | No |
| 211 | ELANE | NA | 13717 | 0.031 | 0.4295 | No |
| 212 | WFDC10A | NA | 13757 | 0.031 | 0.4293 | No |
| 213 | IFNA14 | NA | 14021 | 0.029 | 0.4249 | No |
| 214 | PRTN3 | NA | 14127 | 0.028 | 0.4234 | No |
| 215 | IL17F | NA | 14335 | 0.027 | 0.4201 | No |
| 216 | C8B | NA | 15305 | 0.020 | 0.4025 | No |
| 217 | IL17A | NA | 15583 | 0.018 | 0.3977 | No |
| 218 | IFNA5 | NA | 15607 | 0.018 | 0.3976 | No |
| 219 | F2 | NA | 15633 | 0.018 | 0.3975 | No |
| 220 | HLA-DQB1 | NA | 15698 | 0.018 | 0.3966 | No |
| 221 | CFHR2 | NA | 15845 | 0.017 | 0.3942 | No |
| 222 | SPAG11B | NA | 15971 | 0.016 | 0.3921 | No |
| 223 | IFNA2 | NA | 16038 | 0.016 | 0.3912 | No |
| 224 | IGHA1 | NA | 16252 | 0.015 | 0.3875 | No |
| 225 | IFNA10 | NA | 16334 | 0.014 | 0.3863 | No |
| 226 | IFNA6 | NA | 16469 | 0.013 | 0.3840 | No |
| 227 | HTN3 | NA | 16717 | 0.012 | 0.3797 | No |
| 228 | IFNA16 | NA | 17902 | 0.007 | 0.3579 | No |
| 229 | IFNA13 | NA | 18033 | 0.006 | 0.3556 | No |
| 230 | DEFB126 | NA | 18240 | 0.005 | 0.3519 | No |
| 231 | SPAG11A | NA | 18351 | 0.005 | 0.3500 | No |
| 232 | IFNW1 | NA | 18556 | 0.004 | 0.3463 | No |
| 233 | FCN1 | NA | 18571 | 0.004 | 0.3461 | No |
| 234 | HTN1 | NA | 18582 | 0.004 | 0.3460 | No |
| 235 | DEFA3 | NA | 18608 | 0.004 | 0.3456 | No |
| 236 | CPN1 | NA | 18624 | 0.004 | 0.3454 | No |
| 237 | ZP4 | NA | 18654 | 0.003 | 0.3449 | No |
| 238 | IFNA17 | NA | 18900 | 0.002 | 0.3404 | No |
| 239 | COLEC10 | NA | 18923 | 0.002 | 0.3400 | No |
| 240 | DEFB103A | NA | 19484 | 0.000 | 0.3297 | No |
| 241 | DEFB103B | NA | 22111 | 0.000 | 0.2813 | No |
| 242 | MIR520E | NA | 23439 | 0.000 | 0.2568 | No |
| 243 | MIR520B | NA | 24057 | 0.000 | 0.2454 | No |
| 244 | SPNS2 | NA | 25394 | -0.002 | 0.2208 | No |
| 245 | C7 | NA | 25876 | -0.007 | 0.2121 | No |
| 246 | IGHA2 | NA | 25884 | -0.007 | 0.2121 | No |
| 247 | ATP7A | NA | 25927 | -0.008 | 0.2114 | No |
| 248 | C3 | NA | 26097 | -0.010 | 0.2085 | No |
| 249 | DEFA1 | NA | 26458 | -0.013 | 0.2021 | No |
| 250 | IFNK | NA | 26484 | -0.013 | 0.2018 | No |
| 251 | PHB | NA | 26646 | -0.015 | 0.1991 | No |
| 252 | HLA-DRB1 | NA | 26700 | -0.016 | 0.1984 | No |
| 253 | PGLYRP1 | NA | 27022 | -0.018 | 0.1928 | No |
| 254 | CPN2 | NA | 27129 | -0.019 | 0.1912 | No |
| 255 | HLA-A | NA | 27209 | -0.020 | 0.1901 | No |
| 256 | APCS | NA | 27223 | -0.020 | 0.1902 | No |
| 257 | PGLYRP2 | NA | 27626 | -0.024 | 0.1832 | No |
| 258 | MUC7 | NA | 27733 | -0.025 | 0.1816 | No |
| 259 | MS4A1 | NA | 27738 | -0.025 | 0.1820 | No |
| 260 | REG3A | NA | 28039 | -0.028 | 0.1769 | No |
| 261 | BST1 | NA | 28479 | -0.032 | 0.1693 | No |
| 262 | GATA3 | NA | 28489 | -0.032 | 0.1697 | No |
| 263 | CST9LP1 | NA | 28532 | -0.032 | 0.1695 | No |
| 264 | FCN2 | NA | 28546 | -0.032 | 0.1698 | No |
| 265 | IFNB1 | NA | 28635 | -0.033 | 0.1687 | No |
| 266 | CD83 | NA | 29438 | -0.041 | 0.1546 | No |
| 267 | REG3G | NA | 29761 | -0.044 | 0.1495 | No |
| 268 | DEFB1 | NA | 29780 | -0.045 | 0.1499 | No |
| 269 | WFDC9 | NA | 29831 | -0.045 | 0.1497 | No |
| 270 | BPIFA2 | NA | 29993 | -0.047 | 0.1475 | No |
| 271 | WFDC10B | NA | 30069 | -0.048 | 0.1470 | No |
| 272 | IL7 | NA | 30344 | -0.050 | 0.1427 | No |
| 273 | SH2D1A | NA | 30628 | -0.053 | 0.1384 | No |
| 274 | IFNA8 | NA | 31298 | -0.060 | 0.1271 | No |
| 275 | HRG | NA | 31534 | -0.062 | 0.1238 | No |
| 276 | ANG | NA | 31568 | -0.062 | 0.1243 | No |
| 277 | CFD | NA | 31789 | -0.064 | 0.1213 | No |
| 278 | RNASE7 | NA | 32270 | -0.069 | 0.1136 | No |
| 279 | CR1L | NA | 32281 | -0.069 | 0.1146 | No |
| 280 | CAMP | NA | 32525 | -0.072 | 0.1113 | No |
| 281 | CD55 | NA | 32929 | -0.076 | 0.1052 | No |
| 282 | DMBT1 | NA | 33009 | -0.077 | 0.1050 | No |
| 283 | PSMB10 | NA | 33178 | -0.078 | 0.1032 | No |
| 284 | LCN2 | NA | 33202 | -0.079 | 0.1041 | No |
| 285 | BPIFB2 | NA | 33287 | -0.079 | 0.1039 | No |
| 286 | TNF | NA | 33344 | -0.080 | 0.1042 | No |
| 287 | HLA-E | NA | 33665 | -0.083 | 0.0997 | No |
| 288 | VTN | NA | 34499 | -0.091 | 0.0859 | No |
| 289 | OCIAD1 | NA | 34550 | -0.092 | 0.0865 | No |
| 290 | WFDC11 | NA | 34610 | -0.092 | 0.0870 | No |
| 291 | KLK5 | NA | 34791 | -0.094 | 0.0853 | No |
| 292 | CLU | NA | 35202 | -0.098 | 0.0794 | No |
| 293 | IL1B | NA | 35437 | -0.100 | 0.0768 | No |
| 294 | BCL2 | NA | 35791 | -0.104 | 0.0720 | No |
| 295 | PI3 | NA | 36174 | -0.108 | 0.0668 | No |
| 296 | ZP3 | NA | 36824 | -0.114 | 0.0567 | No |
| 297 | C5AR1 | NA | 37017 | -0.116 | 0.0552 | No |
| 298 | IFNA1 | NA | 37170 | -0.117 | 0.0543 | No |
| 299 | CST9 | NA | 37892 | -0.125 | 0.0431 | No |
| 300 | MBL2 | NA | 38004 | -0.126 | 0.0432 | No |
| 301 | DEFA1B | NA | 38212 | -0.129 | 0.0416 | No |
| 302 | TFE3 | NA | 38443 | -0.131 | 0.0395 | No |
| 303 | PPP2R3C | NA | 38489 | -0.132 | 0.0409 | No |
| 304 | C1RL | NA | 38743 | -0.135 | 0.0386 | No |
| 305 | CCL13 | NA | 38750 | -0.135 | 0.0407 | No |
| 306 | IL6 | NA | 38829 | -0.135 | 0.0416 | No |
| 307 | FOXJ1 | NA | 38999 | -0.137 | 0.0408 | No |
| 308 | NOTCH2 | NA | 39157 | -0.139 | 0.0402 | No |
| 309 | S100A9 | NA | 39514 | -0.143 | 0.0361 | No |
| 310 | IFNG | NA | 39805 | -0.146 | 0.0332 | No |
| 311 | IFNA4 | NA | 39865 | -0.147 | 0.0346 | No |
| 312 | S100A12 | NA | 40126 | -0.150 | 0.0323 | No |
| 313 | DEFB127 | NA | 40641 | -0.156 | 0.0255 | No |
| 314 | TREM1 | NA | 40888 | -0.159 | 0.0236 | No |
| 315 | WFDC12 | NA | 41210 | -0.163 | 0.0205 | No |
| 316 | S100A8 | NA | 41788 | -0.170 | 0.0127 | No |
| 317 | CFB | NA | 42111 | -0.175 | 0.0097 | No |
| 318 | CFI | NA | 42287 | -0.177 | 0.0095 | No |
| 319 | SPINK5 | NA | 42789 | -0.183 | 0.0033 | No |
| 320 | IL36RN | NA | 43192 | -0.188 | -0.0009 | No |
| 321 | CXCL5 | NA | 43233 | -0.189 | 0.0015 | No |
| 322 | COLEC11 | NA | 43431 | -0.192 | 0.0011 | No |
| 323 | PDZD11 | NA | 43514 | -0.193 | 0.0029 | No |
| 324 | WFDC3 | NA | 43722 | -0.196 | 0.0024 | No |
| 325 | KLK3 | NA | 44383 | -0.206 | -0.0063 | No |
| 326 | CXCL8 | NA | 44401 | -0.206 | -0.0032 | No |
| 327 | SUSD4 | NA | 45653 | -0.225 | -0.0225 | No |
| 328 | CST9L | NA | 45726 | -0.226 | -0.0200 | No |
| 329 | RBPJ | NA | 45918 | -0.229 | -0.0196 | No |
| 330 | CXCL2 | NA | 46156 | -0.233 | -0.0201 | No |
| 331 | NOTCH1 | NA | 46598 | -0.240 | -0.0242 | No |
| 332 | TRAF3IP2 | NA | 46693 | -0.242 | -0.0218 | No |
| 333 | WFDC2 | NA | 46744 | -0.243 | -0.0186 | No |
| 334 | BCL3 | NA | 46892 | -0.245 | -0.0172 | No |
| 335 | CXCL6 | NA | 47701 | -0.260 | -0.0277 | No |
| 336 | CXCL3 | NA | 47794 | -0.261 | -0.0250 | No |
| 337 | PPBP | NA | 47910 | -0.264 | -0.0227 | No |
| 338 | GNLY | NA | 48334 | -0.272 | -0.0259 | No |
| 339 | PLA2G6 | NA | 48363 | -0.273 | -0.0218 | No |
| 340 | GALP | NA | 48450 | -0.275 | -0.0188 | No |
| 341 | AZU1 | NA | 48507 | -0.276 | -0.0151 | No |
| 342 | ROMO1 | NA | 48570 | -0.277 | -0.0116 | No |
| 343 | SLC11A1 | NA | 48922 | -0.285 | -0.0133 | No |
| 344 | OCIAD2 | NA | 48984 | -0.286 | -0.0096 | No |
| 345 | C8G | NA | 48988 | -0.286 | -0.0048 | No |
| 346 | CXCL1 | NA | 49333 | -0.294 | -0.0062 | No |
| 347 | CCL2 | NA | 49594 | -0.300 | -0.0059 | No |
| 348 | HMGN2 | NA | 49701 | -0.302 | -0.0027 | No |
| 349 | PF4 | NA | 49702 | -0.302 | 0.0024 | No |
| 350 | CD81 | NA | 49796 | -0.305 | 0.0058 | No |
| 351 | RPL39 | NA | 49811 | -0.305 | 0.0107 | No |
| 352 | PF4V1 | NA | 50355 | -0.318 | 0.0060 | No |
| 353 | HPX | NA | 50642 | -0.326 | 0.0063 | No |
| 354 | CD46 | NA | 50667 | -0.327 | 0.0113 | No |
| 355 | KLK7 | NA | 50750 | -0.329 | 0.0154 | No |
| 356 | RPL30 | NA | 51017 | -0.337 | 0.0161 | No |
| 357 | IFNE | NA | 51596 | -0.354 | 0.0115 | No |
| 358 | TFEB | NA | 51691 | -0.358 | 0.0158 | No |
| 359 | IGHE | NA | 51792 | -0.361 | 0.0200 | No |
| 360 | LEAP2 | NA | 52673 | -0.394 | 0.0104 | No |
| 361 | RARRES2 | NA | 53732 | -0.460 | -0.0013 | No |
| 362 | FAM3A | NA | 54100 | -0.499 | 0.0003 | No |
| 363 | RPS19 | NA | 54218 | -0.515 | 0.0069 | No |