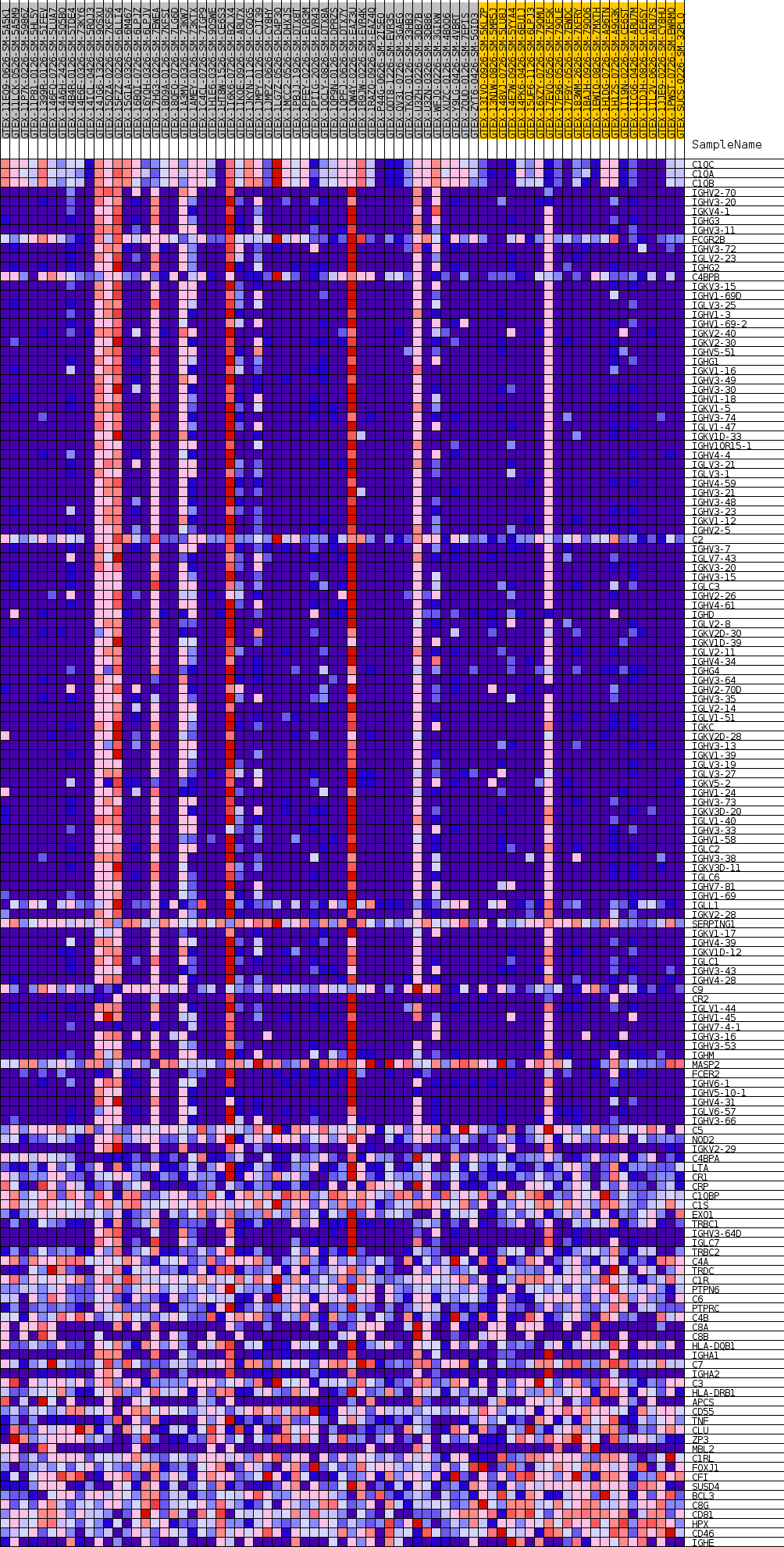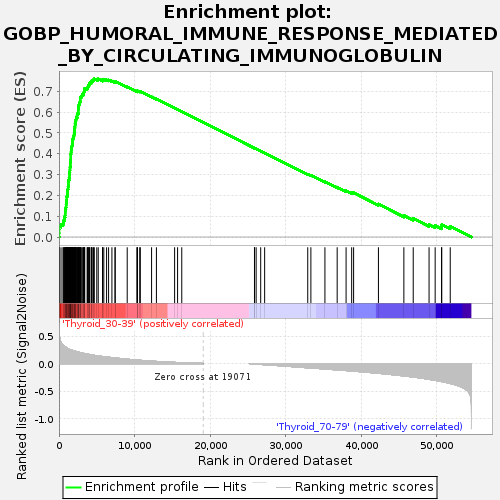
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Thyroid.Thyroid.cls #Thyroid_30-39_versus_Thyroid_70-79.Thyroid.cls #Thyroid_30-39_versus_Thyroid_70-79_repos |
| Phenotype | Thyroid.cls#Thyroid_30-39_versus_Thyroid_70-79_repos |
| Upregulated in class | Thyroid_30-39 |
| GeneSet | GOBP_HUMORAL_IMMUNE_RESPONSE_MEDIATED_BY_CIRCULATING_IMMUNOGLOBULIN |
| Enrichment Score (ES) | 0.75982475 |
| Normalized Enrichment Score (NES) | 1.4109821 |
| Nominal p-value | 0.1384892 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.991 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | C1QC | NA | 22 | 0.551 | 0.0176 | Yes |
| 2 | C1QA | NA | 28 | 0.541 | 0.0352 | Yes |
| 3 | C1QB | NA | 33 | 0.534 | 0.0525 | Yes |
| 4 | IGHV2-70 | NA | 266 | 0.384 | 0.0608 | Yes |
| 5 | IGHV3-20 | NA | 541 | 0.333 | 0.0667 | Yes |
| 6 | IGKV4-1 | NA | 570 | 0.329 | 0.0769 | Yes |
| 7 | IGHG3 | NA | 685 | 0.317 | 0.0851 | Yes |
| 8 | IGHV3-11 | NA | 702 | 0.315 | 0.0951 | Yes |
| 9 | FCGR2B | NA | 799 | 0.305 | 0.1033 | Yes |
| 10 | IGHV3-72 | NA | 821 | 0.303 | 0.1128 | Yes |
| 11 | IGLV2-23 | NA | 825 | 0.303 | 0.1227 | Yes |
| 12 | IGHG2 | NA | 846 | 0.301 | 0.1321 | Yes |
| 13 | C4BPB | NA | 906 | 0.294 | 0.1406 | Yes |
| 14 | IGKV3-15 | NA | 938 | 0.291 | 0.1496 | Yes |
| 15 | IGHV1-69D | NA | 947 | 0.290 | 0.1589 | Yes |
| 16 | IGLV3-25 | NA | 962 | 0.289 | 0.1681 | Yes |
| 17 | IGHV1-3 | NA | 1000 | 0.286 | 0.1767 | Yes |
| 18 | IGHV1-69-2 | NA | 1014 | 0.284 | 0.1858 | Yes |
| 19 | IGKV2-40 | NA | 1015 | 0.284 | 0.1950 | Yes |
| 20 | IGKV2-30 | NA | 1114 | 0.276 | 0.2022 | Yes |
| 21 | IGHV5-51 | NA | 1135 | 0.274 | 0.2108 | Yes |
| 22 | IGHG1 | NA | 1146 | 0.273 | 0.2196 | Yes |
| 23 | IGKV1-16 | NA | 1158 | 0.272 | 0.2283 | Yes |
| 24 | IGHV3-49 | NA | 1199 | 0.270 | 0.2363 | Yes |
| 25 | IGHV3-30 | NA | 1213 | 0.269 | 0.2449 | Yes |
| 26 | IGHV1-18 | NA | 1236 | 0.267 | 0.2532 | Yes |
| 27 | IGKV1-5 | NA | 1241 | 0.267 | 0.2618 | Yes |
| 28 | IGHV3-74 | NA | 1243 | 0.267 | 0.2705 | Yes |
| 29 | IGLV1-47 | NA | 1327 | 0.261 | 0.2775 | Yes |
| 30 | IGKV1D-33 | NA | 1341 | 0.260 | 0.2858 | Yes |
| 31 | IGHV1OR15-1 | NA | 1378 | 0.258 | 0.2935 | Yes |
| 32 | IGHV4-4 | NA | 1379 | 0.258 | 0.3020 | Yes |
| 33 | IGLV3-21 | NA | 1420 | 0.255 | 0.3095 | Yes |
| 34 | IGLV3-1 | NA | 1435 | 0.255 | 0.3176 | Yes |
| 35 | IGHV4-59 | NA | 1451 | 0.254 | 0.3256 | Yes |
| 36 | IGHV3-21 | NA | 1460 | 0.253 | 0.3337 | Yes |
| 37 | IGHV3-48 | NA | 1478 | 0.252 | 0.3417 | Yes |
| 38 | IGHV3-23 | NA | 1510 | 0.251 | 0.3493 | Yes |
| 39 | IGKV1-12 | NA | 1511 | 0.251 | 0.3575 | Yes |
| 40 | IGHV2-5 | NA | 1536 | 0.249 | 0.3652 | Yes |
| 41 | C2 | NA | 1543 | 0.249 | 0.3732 | Yes |
| 42 | IGHV3-7 | NA | 1550 | 0.248 | 0.3812 | Yes |
| 43 | IGLV7-43 | NA | 1552 | 0.248 | 0.3892 | Yes |
| 44 | IGKV3-20 | NA | 1555 | 0.248 | 0.3973 | Yes |
| 45 | IGHV3-15 | NA | 1586 | 0.246 | 0.4048 | Yes |
| 46 | IGLC3 | NA | 1602 | 0.246 | 0.4125 | Yes |
| 47 | IGHV2-26 | NA | 1612 | 0.245 | 0.4204 | Yes |
| 48 | IGHV4-61 | NA | 1690 | 0.242 | 0.4269 | Yes |
| 49 | IGHD | NA | 1697 | 0.241 | 0.4346 | Yes |
| 50 | IGLV2-8 | NA | 1729 | 0.239 | 0.4419 | Yes |
| 51 | IGKV2D-30 | NA | 1768 | 0.238 | 0.4489 | Yes |
| 52 | IGKV1D-39 | NA | 1794 | 0.236 | 0.4562 | Yes |
| 53 | IGLV2-11 | NA | 1811 | 0.236 | 0.4636 | Yes |
| 54 | IGHV4-34 | NA | 1818 | 0.235 | 0.4712 | Yes |
| 55 | IGHG4 | NA | 1903 | 0.232 | 0.4772 | Yes |
| 56 | IGHV3-64 | NA | 1932 | 0.231 | 0.4842 | Yes |
| 57 | IGHV2-70D | NA | 1979 | 0.229 | 0.4908 | Yes |
| 58 | IGHV3-35 | NA | 2011 | 0.228 | 0.4977 | Yes |
| 59 | IGLV2-14 | NA | 2033 | 0.227 | 0.5047 | Yes |
| 60 | IGLV1-51 | NA | 2044 | 0.227 | 0.5119 | Yes |
| 61 | IGKC | NA | 2057 | 0.226 | 0.5191 | Yes |
| 62 | IGKV2D-28 | NA | 2099 | 0.225 | 0.5257 | Yes |
| 63 | IGHV3-13 | NA | 2110 | 0.224 | 0.5328 | Yes |
| 64 | IGKV1-39 | NA | 2126 | 0.224 | 0.5399 | Yes |
| 65 | IGLV3-19 | NA | 2132 | 0.224 | 0.5471 | Yes |
| 66 | IGLV3-27 | NA | 2209 | 0.221 | 0.5529 | Yes |
| 67 | IGKV5-2 | NA | 2231 | 0.220 | 0.5597 | Yes |
| 68 | IGHV1-24 | NA | 2244 | 0.219 | 0.5666 | Yes |
| 69 | IGHV3-73 | NA | 2283 | 0.217 | 0.5730 | Yes |
| 70 | IGKV3D-20 | NA | 2398 | 0.213 | 0.5778 | Yes |
| 71 | IGLV1-40 | NA | 2400 | 0.213 | 0.5848 | Yes |
| 72 | IGHV3-33 | NA | 2419 | 0.212 | 0.5914 | Yes |
| 73 | IGHV1-58 | NA | 2539 | 0.208 | 0.5960 | Yes |
| 74 | IGLC2 | NA | 2547 | 0.208 | 0.6026 | Yes |
| 75 | IGHV3-38 | NA | 2548 | 0.208 | 0.6094 | Yes |
| 76 | IGKV3D-11 | NA | 2573 | 0.207 | 0.6157 | Yes |
| 77 | IGLC6 | NA | 2574 | 0.207 | 0.6225 | Yes |
| 78 | IGHV7-81 | NA | 2622 | 0.205 | 0.6283 | Yes |
| 79 | IGHV1-69 | NA | 2627 | 0.205 | 0.6349 | Yes |
| 80 | IGLL1 | NA | 2697 | 0.202 | 0.6403 | Yes |
| 81 | IGKV2-28 | NA | 2754 | 0.201 | 0.6458 | Yes |
| 82 | SERPING1 | NA | 2784 | 0.199 | 0.6518 | Yes |
| 83 | IGKV1-17 | NA | 2814 | 0.199 | 0.6577 | Yes |
| 84 | IGHV4-39 | NA | 2816 | 0.199 | 0.6642 | Yes |
| 85 | IGKV1D-12 | NA | 2817 | 0.199 | 0.6707 | Yes |
| 86 | IGLC1 | NA | 2899 | 0.195 | 0.6756 | Yes |
| 87 | IGHV3-43 | NA | 3029 | 0.191 | 0.6794 | Yes |
| 88 | IGHV4-28 | NA | 3075 | 0.190 | 0.6848 | Yes |
| 89 | C9 | NA | 3242 | 0.185 | 0.6878 | Yes |
| 90 | CR2 | NA | 3270 | 0.184 | 0.6933 | Yes |
| 91 | IGLV1-44 | NA | 3311 | 0.183 | 0.6985 | Yes |
| 92 | IGHV1-45 | NA | 3364 | 0.182 | 0.7035 | Yes |
| 93 | IGHV7-4-1 | NA | 3393 | 0.181 | 0.7089 | Yes |
| 94 | IGHV3-16 | NA | 3394 | 0.181 | 0.7148 | Yes |
| 95 | IGHV3-53 | NA | 3723 | 0.172 | 0.7144 | Yes |
| 96 | IGHM | NA | 3785 | 0.170 | 0.7188 | Yes |
| 97 | MASP2 | NA | 3857 | 0.168 | 0.7230 | Yes |
| 98 | FCER2 | NA | 3869 | 0.168 | 0.7283 | Yes |
| 99 | IGHV6-1 | NA | 3991 | 0.165 | 0.7314 | Yes |
| 100 | IGHV5-10-1 | NA | 4000 | 0.164 | 0.7366 | Yes |
| 101 | IGHV4-31 | NA | 4069 | 0.163 | 0.7407 | Yes |
| 102 | IGLV6-57 | NA | 4269 | 0.158 | 0.7422 | Yes |
| 103 | IGHV3-66 | NA | 4327 | 0.156 | 0.7462 | Yes |
| 104 | C5 | NA | 4330 | 0.156 | 0.7513 | Yes |
| 105 | NOD2 | NA | 4498 | 0.152 | 0.7532 | Yes |
| 106 | IGKV2-29 | NA | 4573 | 0.151 | 0.7568 | Yes |
| 107 | C4BPA | NA | 4672 | 0.149 | 0.7598 | Yes |
| 108 | LTA | NA | 5001 | 0.142 | 0.7584 | No |
| 109 | CR1 | NA | 5194 | 0.139 | 0.7594 | No |
| 110 | CRP | NA | 5763 | 0.127 | 0.7532 | No |
| 111 | C1QBP | NA | 5767 | 0.127 | 0.7573 | No |
| 112 | C1S | NA | 5931 | 0.124 | 0.7583 | No |
| 113 | EXO1 | NA | 6298 | 0.118 | 0.7555 | No |
| 114 | TRBC1 | NA | 6565 | 0.114 | 0.7543 | No |
| 115 | IGHV3-64D | NA | 7005 | 0.107 | 0.7497 | No |
| 116 | IGLC7 | NA | 7419 | 0.100 | 0.7454 | No |
| 117 | TRBC2 | NA | 7436 | 0.100 | 0.7484 | No |
| 118 | C4A | NA | 9029 | 0.078 | 0.7217 | No |
| 119 | TRDC | NA | 10333 | 0.062 | 0.6998 | No |
| 120 | C1R | NA | 10343 | 0.062 | 0.7016 | No |
| 121 | PTPN6 | NA | 10393 | 0.062 | 0.7027 | No |
| 122 | C6 | NA | 10692 | 0.058 | 0.6992 | No |
| 123 | PTPRC | NA | 10760 | 0.058 | 0.6998 | No |
| 124 | C4B | NA | 12255 | 0.043 | 0.6738 | No |
| 125 | C8A | NA | 12895 | 0.038 | 0.6633 | No |
| 126 | C8B | NA | 15305 | 0.020 | 0.6197 | No |
| 127 | HLA-DQB1 | NA | 15698 | 0.018 | 0.6130 | No |
| 128 | IGHA1 | NA | 16252 | 0.015 | 0.6034 | No |
| 129 | C7 | NA | 25876 | -0.007 | 0.4268 | No |
| 130 | IGHA2 | NA | 25884 | -0.007 | 0.4270 | No |
| 131 | C3 | NA | 26097 | -0.010 | 0.4234 | No |
| 132 | HLA-DRB1 | NA | 26700 | -0.016 | 0.4128 | No |
| 133 | APCS | NA | 27223 | -0.020 | 0.4039 | No |
| 134 | CD55 | NA | 32929 | -0.076 | 0.3016 | No |
| 135 | TNF | NA | 33344 | -0.080 | 0.2966 | No |
| 136 | CLU | NA | 35202 | -0.098 | 0.2657 | No |
| 137 | ZP3 | NA | 36824 | -0.114 | 0.2397 | No |
| 138 | MBL2 | NA | 38004 | -0.126 | 0.2221 | No |
| 139 | C1RL | NA | 38743 | -0.135 | 0.2130 | No |
| 140 | FOXJ1 | NA | 38999 | -0.137 | 0.2127 | No |
| 141 | CFI | NA | 42287 | -0.177 | 0.1581 | No |
| 142 | SUSD4 | NA | 45653 | -0.225 | 0.1037 | No |
| 143 | BCL3 | NA | 46892 | -0.245 | 0.0889 | No |
| 144 | C8G | NA | 48988 | -0.286 | 0.0598 | No |
| 145 | CD81 | NA | 49796 | -0.305 | 0.0549 | No |
| 146 | HPX | NA | 50642 | -0.326 | 0.0500 | No |
| 147 | CD46 | NA | 50667 | -0.327 | 0.0603 | No |
| 148 | IGHE | NA | 51792 | -0.361 | 0.0514 | No |