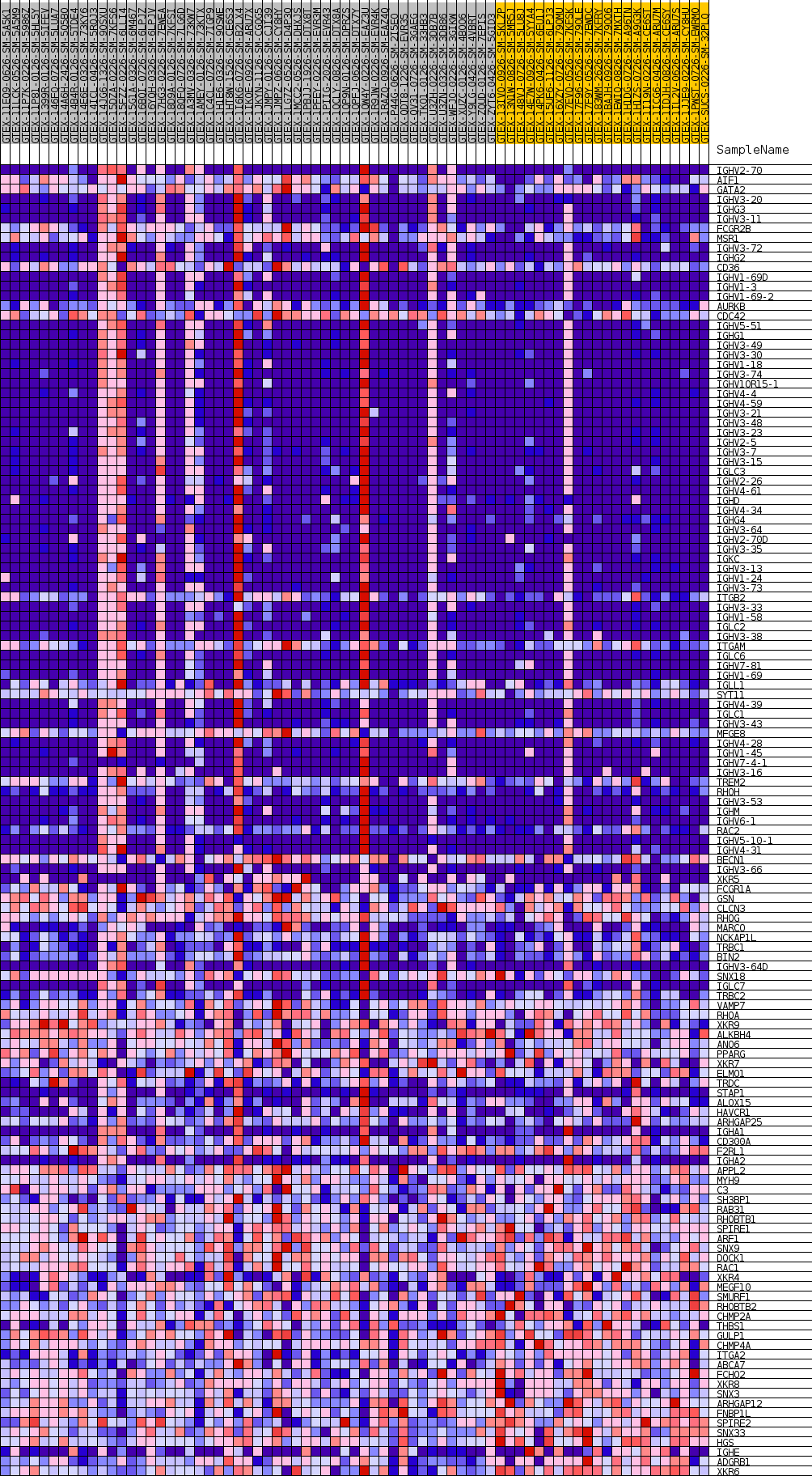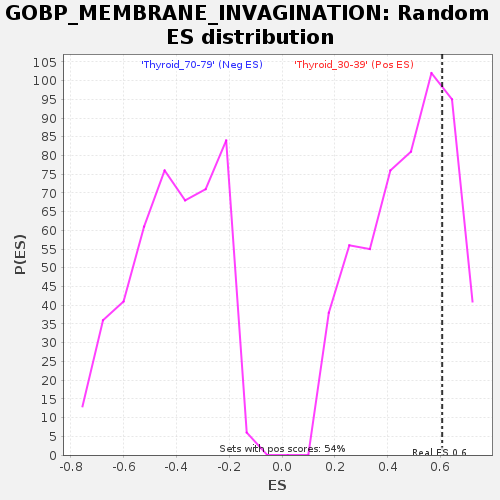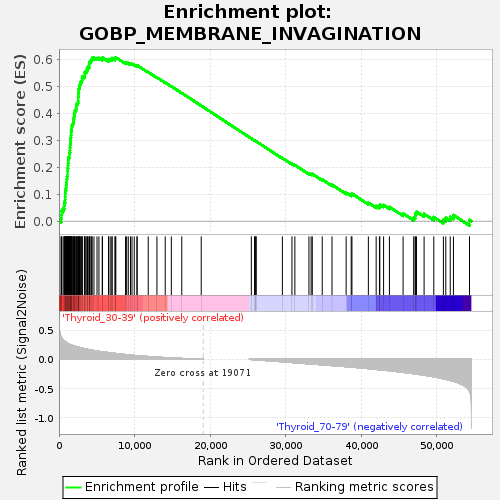
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Thyroid.Thyroid.cls #Thyroid_30-39_versus_Thyroid_70-79.Thyroid.cls #Thyroid_30-39_versus_Thyroid_70-79_repos |
| Phenotype | Thyroid.cls#Thyroid_30-39_versus_Thyroid_70-79_repos |
| Upregulated in class | Thyroid_30-39 |
| GeneSet | GOBP_MEMBRANE_INVAGINATION |
| Enrichment Score (ES) | 0.60687226 |
| Normalized Enrichment Score (NES) | 1.2726007 |
| Nominal p-value | 0.24632353 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | IGHV2-70 | NA | 266 | 0.384 | 0.0101 | Yes |
| 2 | AIF1 | NA | 334 | 0.368 | 0.0233 | Yes |
| 3 | GATA2 | NA | 348 | 0.365 | 0.0373 | Yes |
| 4 | IGHV3-20 | NA | 541 | 0.333 | 0.0467 | Yes |
| 5 | IGHG3 | NA | 685 | 0.317 | 0.0565 | Yes |
| 6 | IGHV3-11 | NA | 702 | 0.315 | 0.0685 | Yes |
| 7 | FCGR2B | NA | 799 | 0.305 | 0.0786 | Yes |
| 8 | MSR1 | NA | 812 | 0.304 | 0.0903 | Yes |
| 9 | IGHV3-72 | NA | 821 | 0.303 | 0.1020 | Yes |
| 10 | IGHG2 | NA | 846 | 0.301 | 0.1133 | Yes |
| 11 | CD36 | NA | 926 | 0.292 | 0.1232 | Yes |
| 12 | IGHV1-69D | NA | 947 | 0.290 | 0.1342 | Yes |
| 13 | IGHV1-3 | NA | 1000 | 0.286 | 0.1444 | Yes |
| 14 | IGHV1-69-2 | NA | 1014 | 0.284 | 0.1552 | Yes |
| 15 | AURKB | NA | 1061 | 0.280 | 0.1653 | Yes |
| 16 | CDC42 | NA | 1134 | 0.274 | 0.1747 | Yes |
| 17 | IGHV5-51 | NA | 1135 | 0.274 | 0.1854 | Yes |
| 18 | IGHG1 | NA | 1146 | 0.273 | 0.1959 | Yes |
| 19 | IGHV3-49 | NA | 1199 | 0.270 | 0.2055 | Yes |
| 20 | IGHV3-30 | NA | 1213 | 0.269 | 0.2158 | Yes |
| 21 | IGHV1-18 | NA | 1236 | 0.267 | 0.2258 | Yes |
| 22 | IGHV3-74 | NA | 1243 | 0.267 | 0.2361 | Yes |
| 23 | IGHV1OR15-1 | NA | 1378 | 0.258 | 0.2437 | Yes |
| 24 | IGHV4-4 | NA | 1379 | 0.258 | 0.2538 | Yes |
| 25 | IGHV4-59 | NA | 1451 | 0.254 | 0.2624 | Yes |
| 26 | IGHV3-21 | NA | 1460 | 0.253 | 0.2721 | Yes |
| 27 | IGHV3-48 | NA | 1478 | 0.252 | 0.2816 | Yes |
| 28 | IGHV3-23 | NA | 1510 | 0.251 | 0.2909 | Yes |
| 29 | IGHV2-5 | NA | 1536 | 0.249 | 0.3001 | Yes |
| 30 | IGHV3-7 | NA | 1550 | 0.248 | 0.3096 | Yes |
| 31 | IGHV3-15 | NA | 1586 | 0.246 | 0.3185 | Yes |
| 32 | IGLC3 | NA | 1602 | 0.246 | 0.3279 | Yes |
| 33 | IGHV2-26 | NA | 1612 | 0.245 | 0.3373 | Yes |
| 34 | IGHV4-61 | NA | 1690 | 0.242 | 0.3453 | Yes |
| 35 | IGHD | NA | 1697 | 0.241 | 0.3546 | Yes |
| 36 | IGHV4-34 | NA | 1818 | 0.235 | 0.3616 | Yes |
| 37 | IGHG4 | NA | 1903 | 0.232 | 0.3691 | Yes |
| 38 | IGHV3-64 | NA | 1932 | 0.231 | 0.3776 | Yes |
| 39 | IGHV2-70D | NA | 1979 | 0.229 | 0.3856 | Yes |
| 40 | IGHV3-35 | NA | 2011 | 0.228 | 0.3940 | Yes |
| 41 | IGKC | NA | 2057 | 0.226 | 0.4020 | Yes |
| 42 | IGHV3-13 | NA | 2110 | 0.224 | 0.4098 | Yes |
| 43 | IGHV1-24 | NA | 2244 | 0.219 | 0.4159 | Yes |
| 44 | IGHV3-73 | NA | 2283 | 0.217 | 0.4237 | Yes |
| 45 | ITGB2 | NA | 2287 | 0.217 | 0.4321 | Yes |
| 46 | IGHV3-33 | NA | 2419 | 0.212 | 0.4380 | Yes |
| 47 | IGHV1-58 | NA | 2539 | 0.208 | 0.4439 | Yes |
| 48 | IGLC2 | NA | 2547 | 0.208 | 0.4519 | Yes |
| 49 | IGHV3-38 | NA | 2548 | 0.208 | 0.4600 | Yes |
| 50 | ITGAM | NA | 2572 | 0.207 | 0.4677 | Yes |
| 51 | IGLC6 | NA | 2574 | 0.207 | 0.4757 | Yes |
| 52 | IGHV7-81 | NA | 2622 | 0.205 | 0.4829 | Yes |
| 53 | IGHV1-69 | NA | 2627 | 0.205 | 0.4908 | Yes |
| 54 | IGLL1 | NA | 2697 | 0.202 | 0.4974 | Yes |
| 55 | SYT11 | NA | 2726 | 0.201 | 0.5048 | Yes |
| 56 | IGHV4-39 | NA | 2816 | 0.199 | 0.5109 | Yes |
| 57 | IGLC1 | NA | 2899 | 0.195 | 0.5170 | Yes |
| 58 | IGHV3-43 | NA | 3029 | 0.191 | 0.5221 | Yes |
| 59 | MFGE8 | NA | 3057 | 0.190 | 0.5290 | Yes |
| 60 | IGHV4-28 | NA | 3075 | 0.190 | 0.5361 | Yes |
| 61 | IGHV1-45 | NA | 3364 | 0.182 | 0.5379 | Yes |
| 62 | IGHV7-4-1 | NA | 3393 | 0.181 | 0.5445 | Yes |
| 63 | IGHV3-16 | NA | 3394 | 0.181 | 0.5515 | Yes |
| 64 | TREM2 | NA | 3542 | 0.176 | 0.5557 | Yes |
| 65 | RHOH | NA | 3709 | 0.172 | 0.5594 | Yes |
| 66 | IGHV3-53 | NA | 3723 | 0.172 | 0.5659 | Yes |
| 67 | IGHM | NA | 3785 | 0.170 | 0.5714 | Yes |
| 68 | IGHV6-1 | NA | 3991 | 0.165 | 0.5740 | Yes |
| 69 | RAC2 | NA | 3993 | 0.165 | 0.5804 | Yes |
| 70 | IGHV5-10-1 | NA | 4000 | 0.164 | 0.5867 | Yes |
| 71 | IGHV4-31 | NA | 4069 | 0.163 | 0.5918 | Yes |
| 72 | BECN1 | NA | 4229 | 0.159 | 0.5951 | Yes |
| 73 | IGHV3-66 | NA | 4327 | 0.156 | 0.5994 | Yes |
| 74 | XKR5 | NA | 4383 | 0.155 | 0.6045 | Yes |
| 75 | FCGR1A | NA | 4630 | 0.149 | 0.6058 | Yes |
| 76 | GSN | NA | 5031 | 0.142 | 0.6040 | Yes |
| 77 | CLCN3 | NA | 5285 | 0.137 | 0.6046 | Yes |
| 78 | RHOG | NA | 5724 | 0.128 | 0.6016 | Yes |
| 79 | MARCO | NA | 5754 | 0.127 | 0.6060 | Yes |
| 80 | NCKAP1L | NA | 6555 | 0.114 | 0.5958 | Yes |
| 81 | TRBC1 | NA | 6565 | 0.114 | 0.6001 | Yes |
| 82 | BIN2 | NA | 6773 | 0.110 | 0.6006 | Yes |
| 83 | IGHV3-64D | NA | 7005 | 0.107 | 0.6005 | Yes |
| 84 | SNX18 | NA | 7030 | 0.106 | 0.6042 | Yes |
| 85 | IGLC7 | NA | 7419 | 0.100 | 0.6010 | Yes |
| 86 | TRBC2 | NA | 7436 | 0.100 | 0.6046 | Yes |
| 87 | VAMP7 | NA | 7522 | 0.098 | 0.6069 | Yes |
| 88 | RHOA | NA | 8808 | 0.080 | 0.5864 | No |
| 89 | XKR9 | NA | 8955 | 0.079 | 0.5868 | No |
| 90 | ALKBH4 | NA | 9171 | 0.076 | 0.5858 | No |
| 91 | ANO6 | NA | 9465 | 0.073 | 0.5833 | No |
| 92 | PPARG | NA | 9664 | 0.070 | 0.5824 | No |
| 93 | XKR7 | NA | 9960 | 0.066 | 0.5796 | No |
| 94 | ELMO1 | NA | 10332 | 0.062 | 0.5752 | No |
| 95 | TRDC | NA | 10333 | 0.062 | 0.5776 | No |
| 96 | STAP1 | NA | 11812 | 0.047 | 0.5523 | No |
| 97 | ALOX15 | NA | 12964 | 0.037 | 0.5326 | No |
| 98 | HAVCR1 | NA | 14062 | 0.029 | 0.5136 | No |
| 99 | ARHGAP25 | NA | 14870 | 0.023 | 0.4997 | No |
| 100 | IGHA1 | NA | 16252 | 0.015 | 0.4749 | No |
| 101 | CD300A | NA | 18827 | 0.002 | 0.4277 | No |
| 102 | F2RL1 | NA | 25460 | -0.003 | 0.3060 | No |
| 103 | IGHA2 | NA | 25884 | -0.007 | 0.2985 | No |
| 104 | APPL2 | NA | 25968 | -0.009 | 0.2974 | No |
| 105 | MYH9 | NA | 26057 | -0.009 | 0.2961 | No |
| 106 | C3 | NA | 26097 | -0.010 | 0.2958 | No |
| 107 | SH3BP1 | NA | 29570 | -0.042 | 0.2337 | No |
| 108 | RAB31 | NA | 30838 | -0.055 | 0.2125 | No |
| 109 | RHOBTB1 | NA | 31223 | -0.059 | 0.2078 | No |
| 110 | SPIRE1 | NA | 33086 | -0.077 | 0.1766 | No |
| 111 | ARF1 | NA | 33376 | -0.080 | 0.1744 | No |
| 112 | SNX9 | NA | 33531 | -0.082 | 0.1748 | No |
| 113 | DOCK1 | NA | 34846 | -0.095 | 0.1544 | No |
| 114 | RAC1 | NA | 36138 | -0.107 | 0.1349 | No |
| 115 | XKR4 | NA | 38011 | -0.126 | 0.1054 | No |
| 116 | MEGF10 | NA | 38681 | -0.134 | 0.0983 | No |
| 117 | SMURF1 | NA | 38779 | -0.135 | 0.1018 | No |
| 118 | RHOBTB2 | NA | 40954 | -0.160 | 0.0681 | No |
| 119 | CHMP2A | NA | 41986 | -0.173 | 0.0560 | No |
| 120 | THBS1 | NA | 42420 | -0.179 | 0.0550 | No |
| 121 | GULP1 | NA | 42469 | -0.179 | 0.0611 | No |
| 122 | CHMP4A | NA | 42968 | -0.186 | 0.0592 | No |
| 123 | ITGA2 | NA | 43741 | -0.196 | 0.0527 | No |
| 124 | ABCA7 | NA | 45549 | -0.223 | 0.0282 | No |
| 125 | FCHO2 | NA | 46951 | -0.247 | 0.0121 | No |
| 126 | XKR8 | NA | 47147 | -0.250 | 0.0183 | No |
| 127 | SNX3 | NA | 47176 | -0.250 | 0.0275 | No |
| 128 | ARHGAP12 | NA | 47323 | -0.253 | 0.0347 | No |
| 129 | FNBP1L | NA | 48331 | -0.272 | 0.0268 | No |
| 130 | SPIRE2 | NA | 49614 | -0.300 | 0.0150 | No |
| 131 | SNX33 | NA | 50885 | -0.333 | 0.0047 | No |
| 132 | HGS | NA | 51203 | -0.343 | 0.0122 | No |
| 133 | IGHE | NA | 51792 | -0.361 | 0.0155 | No |
| 134 | ADGRB1 | NA | 52214 | -0.377 | 0.0225 | No |
| 135 | XKR6 | NA | 54342 | -0.542 | 0.0046 | No |