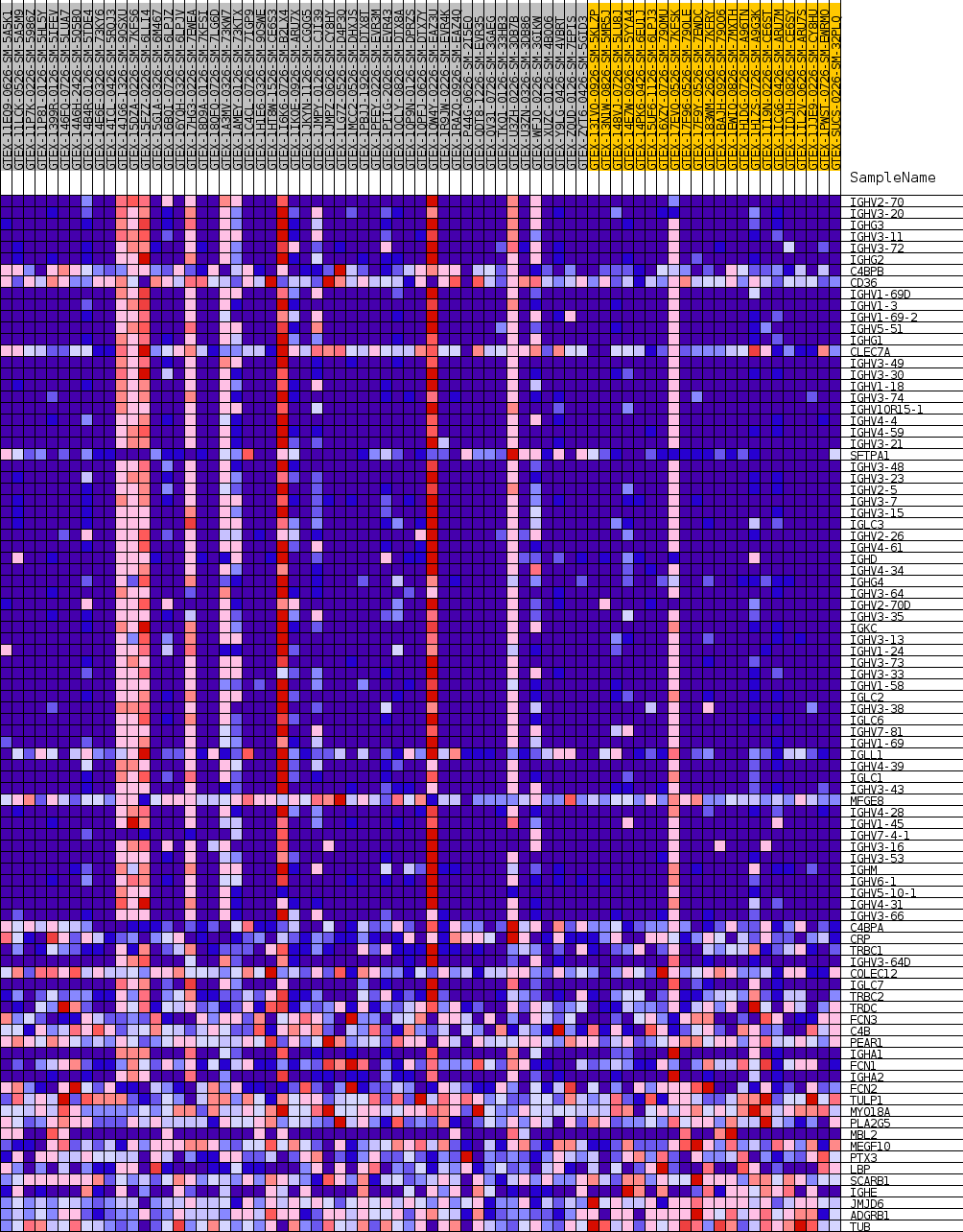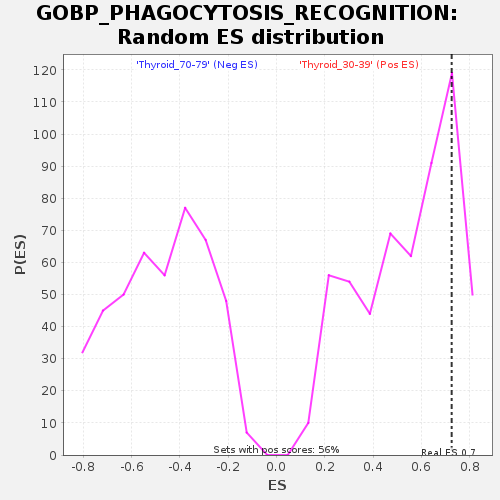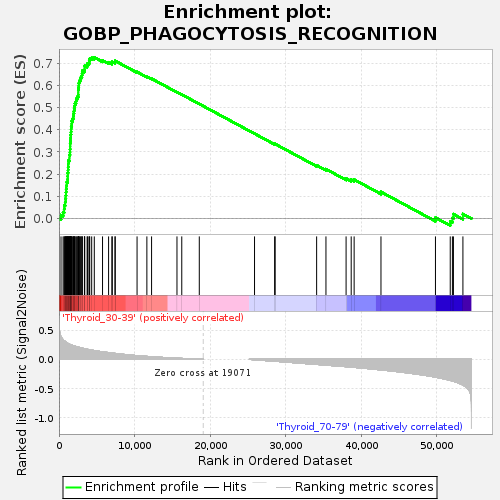
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Thyroid.Thyroid.cls #Thyroid_30-39_versus_Thyroid_70-79.Thyroid.cls #Thyroid_30-39_versus_Thyroid_70-79_repos |
| Phenotype | Thyroid.cls#Thyroid_30-39_versus_Thyroid_70-79_repos |
| Upregulated in class | Thyroid_30-39 |
| GeneSet | GOBP_PHAGOCYTOSIS_RECOGNITION |
| Enrichment Score (ES) | 0.72600704 |
| Normalized Enrichment Score (NES) | 1.3430126 |
| Nominal p-value | 0.19459459 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.997 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | IGHV2-70 | NA | 266 | 0.384 | 0.0157 | Yes |
| 2 | IGHV3-20 | NA | 541 | 0.333 | 0.0285 | Yes |
| 3 | IGHG3 | NA | 685 | 0.317 | 0.0429 | Yes |
| 4 | IGHV3-11 | NA | 702 | 0.315 | 0.0594 | Yes |
| 5 | IGHV3-72 | NA | 821 | 0.303 | 0.0735 | Yes |
| 6 | IGHG2 | NA | 846 | 0.301 | 0.0892 | Yes |
| 7 | C4BPB | NA | 906 | 0.294 | 0.1039 | Yes |
| 8 | CD36 | NA | 926 | 0.292 | 0.1192 | Yes |
| 9 | IGHV1-69D | NA | 947 | 0.290 | 0.1344 | Yes |
| 10 | IGHV1-3 | NA | 1000 | 0.286 | 0.1487 | Yes |
| 11 | IGHV1-69-2 | NA | 1014 | 0.284 | 0.1637 | Yes |
| 12 | IGHV5-51 | NA | 1135 | 0.274 | 0.1762 | Yes |
| 13 | IGHG1 | NA | 1146 | 0.273 | 0.1906 | Yes |
| 14 | CLEC7A | NA | 1154 | 0.273 | 0.2051 | Yes |
| 15 | IGHV3-49 | NA | 1199 | 0.270 | 0.2188 | Yes |
| 16 | IGHV3-30 | NA | 1213 | 0.269 | 0.2330 | Yes |
| 17 | IGHV1-18 | NA | 1236 | 0.267 | 0.2469 | Yes |
| 18 | IGHV3-74 | NA | 1243 | 0.267 | 0.2610 | Yes |
| 19 | IGHV1OR15-1 | NA | 1378 | 0.258 | 0.2724 | Yes |
| 20 | IGHV4-4 | NA | 1379 | 0.258 | 0.2862 | Yes |
| 21 | IGHV4-59 | NA | 1451 | 0.254 | 0.2985 | Yes |
| 22 | IGHV3-21 | NA | 1460 | 0.253 | 0.3119 | Yes |
| 23 | SFTPA1 | NA | 1475 | 0.252 | 0.3252 | Yes |
| 24 | IGHV3-48 | NA | 1478 | 0.252 | 0.3387 | Yes |
| 25 | IGHV3-23 | NA | 1510 | 0.251 | 0.3515 | Yes |
| 26 | IGHV2-5 | NA | 1536 | 0.249 | 0.3644 | Yes |
| 27 | IGHV3-7 | NA | 1550 | 0.248 | 0.3775 | Yes |
| 28 | IGHV3-15 | NA | 1586 | 0.246 | 0.3900 | Yes |
| 29 | IGLC3 | NA | 1602 | 0.246 | 0.4029 | Yes |
| 30 | IGHV2-26 | NA | 1612 | 0.245 | 0.4159 | Yes |
| 31 | IGHV4-61 | NA | 1690 | 0.242 | 0.4274 | Yes |
| 32 | IGHD | NA | 1697 | 0.241 | 0.4402 | Yes |
| 33 | IGHV4-34 | NA | 1818 | 0.235 | 0.4506 | Yes |
| 34 | IGHG4 | NA | 1903 | 0.232 | 0.4615 | Yes |
| 35 | IGHV3-64 | NA | 1932 | 0.231 | 0.4733 | Yes |
| 36 | IGHV2-70D | NA | 1979 | 0.229 | 0.4848 | Yes |
| 37 | IGHV3-35 | NA | 2011 | 0.228 | 0.4964 | Yes |
| 38 | IGKC | NA | 2057 | 0.226 | 0.5077 | Yes |
| 39 | IGHV3-13 | NA | 2110 | 0.224 | 0.5188 | Yes |
| 40 | IGHV1-24 | NA | 2244 | 0.219 | 0.5281 | Yes |
| 41 | IGHV3-73 | NA | 2283 | 0.217 | 0.5390 | Yes |
| 42 | IGHV3-33 | NA | 2419 | 0.212 | 0.5479 | Yes |
| 43 | IGHV1-58 | NA | 2539 | 0.208 | 0.5569 | Yes |
| 44 | IGLC2 | NA | 2547 | 0.208 | 0.5679 | Yes |
| 45 | IGHV3-38 | NA | 2548 | 0.208 | 0.5790 | Yes |
| 46 | IGLC6 | NA | 2574 | 0.207 | 0.5896 | Yes |
| 47 | IGHV7-81 | NA | 2622 | 0.205 | 0.5997 | Yes |
| 48 | IGHV1-69 | NA | 2627 | 0.205 | 0.6106 | Yes |
| 49 | IGLL1 | NA | 2697 | 0.202 | 0.6202 | Yes |
| 50 | IGHV4-39 | NA | 2816 | 0.199 | 0.6287 | Yes |
| 51 | IGLC1 | NA | 2899 | 0.195 | 0.6376 | Yes |
| 52 | IGHV3-43 | NA | 3029 | 0.191 | 0.6455 | Yes |
| 53 | MFGE8 | NA | 3057 | 0.190 | 0.6552 | Yes |
| 54 | IGHV4-28 | NA | 3075 | 0.190 | 0.6651 | Yes |
| 55 | IGHV1-45 | NA | 3364 | 0.182 | 0.6695 | Yes |
| 56 | IGHV7-4-1 | NA | 3393 | 0.181 | 0.6787 | Yes |
| 57 | IGHV3-16 | NA | 3394 | 0.181 | 0.6884 | Yes |
| 58 | IGHV3-53 | NA | 3723 | 0.172 | 0.6916 | Yes |
| 59 | IGHM | NA | 3785 | 0.170 | 0.6995 | Yes |
| 60 | IGHV6-1 | NA | 3991 | 0.165 | 0.7046 | Yes |
| 61 | IGHV5-10-1 | NA | 4000 | 0.164 | 0.7133 | Yes |
| 62 | IGHV4-31 | NA | 4069 | 0.163 | 0.7207 | Yes |
| 63 | IGHV3-66 | NA | 4327 | 0.156 | 0.7244 | Yes |
| 64 | C4BPA | NA | 4672 | 0.149 | 0.7260 | Yes |
| 65 | CRP | NA | 5763 | 0.127 | 0.7128 | No |
| 66 | TRBC1 | NA | 6565 | 0.114 | 0.7042 | No |
| 67 | IGHV3-64D | NA | 7005 | 0.107 | 0.7019 | No |
| 68 | COLEC12 | NA | 7051 | 0.106 | 0.7067 | No |
| 69 | IGLC7 | NA | 7419 | 0.100 | 0.7054 | No |
| 70 | TRBC2 | NA | 7436 | 0.100 | 0.7104 | No |
| 71 | TRDC | NA | 10333 | 0.062 | 0.6606 | No |
| 72 | FCN3 | NA | 11632 | 0.049 | 0.6394 | No |
| 73 | C4B | NA | 12255 | 0.043 | 0.6303 | No |
| 74 | PEAR1 | NA | 15617 | 0.018 | 0.5696 | No |
| 75 | IGHA1 | NA | 16252 | 0.015 | 0.5587 | No |
| 76 | FCN1 | NA | 18571 | 0.004 | 0.5164 | No |
| 77 | IGHA2 | NA | 25884 | -0.007 | 0.3827 | No |
| 78 | FCN2 | NA | 28546 | -0.032 | 0.3356 | No |
| 79 | TULP1 | NA | 28612 | -0.033 | 0.3361 | No |
| 80 | MYO18A | NA | 34105 | -0.087 | 0.2400 | No |
| 81 | PLA2G5 | NA | 35339 | -0.099 | 0.2227 | No |
| 82 | MBL2 | NA | 38004 | -0.126 | 0.1806 | No |
| 83 | MEGF10 | NA | 38681 | -0.134 | 0.1754 | No |
| 84 | PTX3 | NA | 39070 | -0.138 | 0.1756 | No |
| 85 | LBP | NA | 42622 | -0.181 | 0.1202 | No |
| 86 | SCARB1 | NA | 49835 | -0.306 | 0.0042 | No |
| 87 | IGHE | NA | 51792 | -0.361 | -0.0123 | No |
| 88 | JMJD6 | NA | 52080 | -0.371 | 0.0023 | No |
| 89 | ADGRB1 | NA | 52214 | -0.377 | 0.0200 | No |
| 90 | TUB | NA | 53470 | -0.440 | 0.0206 | No |