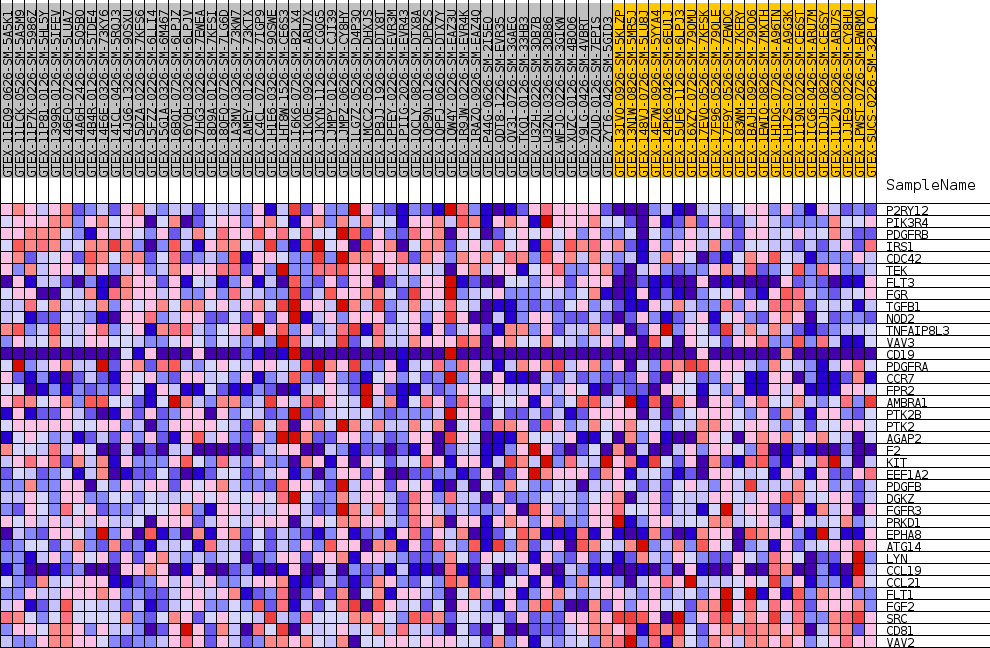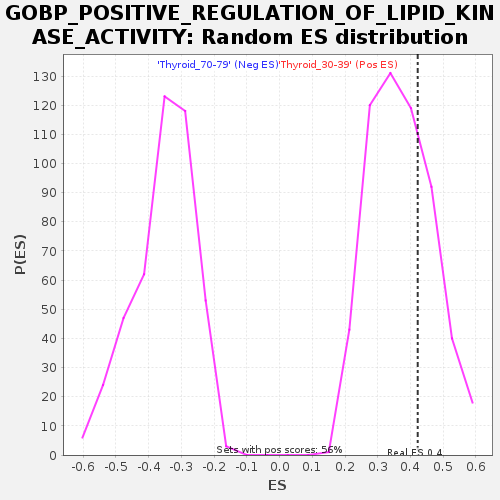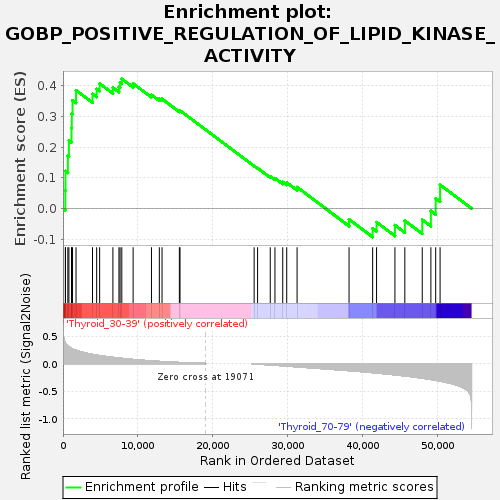
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Thyroid.Thyroid.cls #Thyroid_30-39_versus_Thyroid_70-79.Thyroid.cls #Thyroid_30-39_versus_Thyroid_70-79_repos |
| Phenotype | Thyroid.cls#Thyroid_30-39_versus_Thyroid_70-79_repos |
| Upregulated in class | Thyroid_30-39 |
| GeneSet | GOBP_POSITIVE_REGULATION_OF_LIPID_KINASE_ACTIVITY |
| Enrichment Score (ES) | 0.42215836 |
| Normalized Enrichment Score (NES) | 1.136847 |
| Nominal p-value | 0.30851063 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | P2RY12 | NA | 315 | 0.372 | 0.0584 | Yes |
| 2 | PIK3R4 | NA | 330 | 0.369 | 0.1218 | Yes |
| 3 | PDGFRB | NA | 647 | 0.321 | 0.1713 | Yes |
| 4 | IRS1 | NA | 787 | 0.306 | 0.2215 | Yes |
| 5 | CDC42 | NA | 1134 | 0.274 | 0.2625 | Yes |
| 6 | TEK | NA | 1169 | 0.272 | 0.3087 | Yes |
| 7 | FLT3 | NA | 1273 | 0.265 | 0.3524 | Yes |
| 8 | FGR | NA | 1741 | 0.239 | 0.3850 | Yes |
| 9 | TGFB1 | NA | 3942 | 0.166 | 0.3734 | Yes |
| 10 | NOD2 | NA | 4498 | 0.152 | 0.3895 | Yes |
| 11 | TNFAIP8L3 | NA | 4897 | 0.144 | 0.4070 | Yes |
| 12 | VAV3 | NA | 6667 | 0.112 | 0.3939 | Yes |
| 13 | CD19 | NA | 7476 | 0.099 | 0.3962 | Yes |
| 14 | PDGFRA | NA | 7626 | 0.097 | 0.4102 | Yes |
| 15 | CCR7 | NA | 7857 | 0.094 | 0.4222 | Yes |
| 16 | FPR2 | NA | 9369 | 0.074 | 0.4072 | No |
| 17 | AMBRA1 | NA | 11821 | 0.047 | 0.3703 | No |
| 18 | PTK2B | NA | 12870 | 0.038 | 0.3576 | No |
| 19 | PTK2 | NA | 13232 | 0.035 | 0.3570 | No |
| 20 | AGAP2 | NA | 15549 | 0.018 | 0.3178 | No |
| 21 | F2 | NA | 15633 | 0.018 | 0.3193 | No |
| 22 | KIT | NA | 25538 | -0.004 | 0.1384 | No |
| 23 | EEF1A2 | NA | 25999 | -0.009 | 0.1315 | No |
| 24 | PDGFB | NA | 27693 | -0.025 | 0.1048 | No |
| 25 | DGKZ | NA | 28319 | -0.030 | 0.0985 | No |
| 26 | FGFR3 | NA | 29360 | -0.040 | 0.0864 | No |
| 27 | PRKD1 | NA | 29891 | -0.046 | 0.0846 | No |
| 28 | EPHA8 | NA | 31276 | -0.060 | 0.0695 | No |
| 29 | ATG14 | NA | 38219 | -0.129 | -0.0356 | No |
| 30 | LYN | NA | 41372 | -0.165 | -0.0649 | No |
| 31 | CCL19 | NA | 41896 | -0.172 | -0.0449 | No |
| 32 | CCL21 | NA | 44349 | -0.205 | -0.0544 | No |
| 33 | FLT1 | NA | 45673 | -0.225 | -0.0399 | No |
| 34 | FGF2 | NA | 48000 | -0.265 | -0.0368 | No |
| 35 | SRC | NA | 49154 | -0.290 | -0.0080 | No |
| 36 | CD81 | NA | 49796 | -0.305 | 0.0328 | No |
| 37 | VAV2 | NA | 50383 | -0.319 | 0.0771 | No |