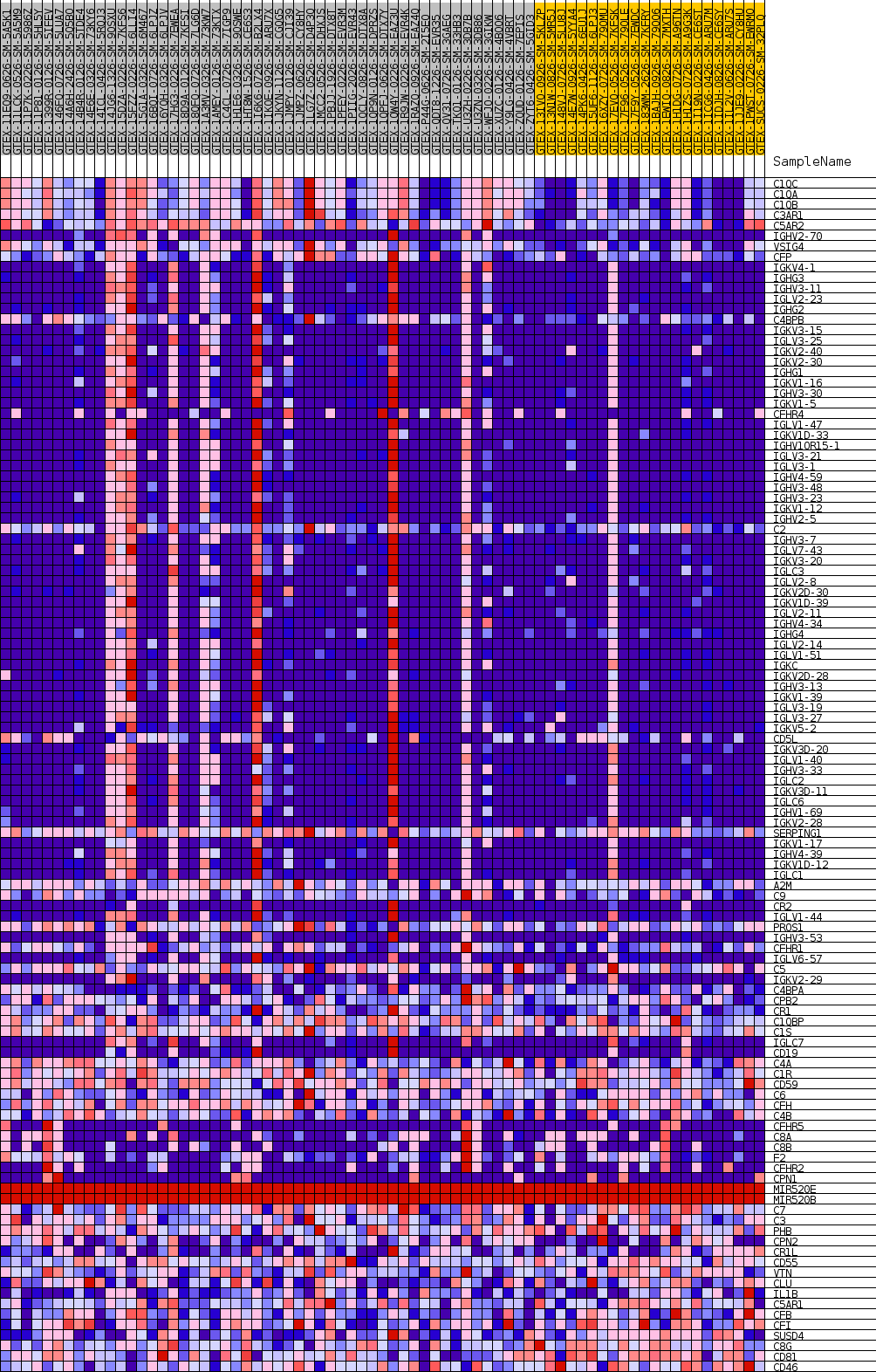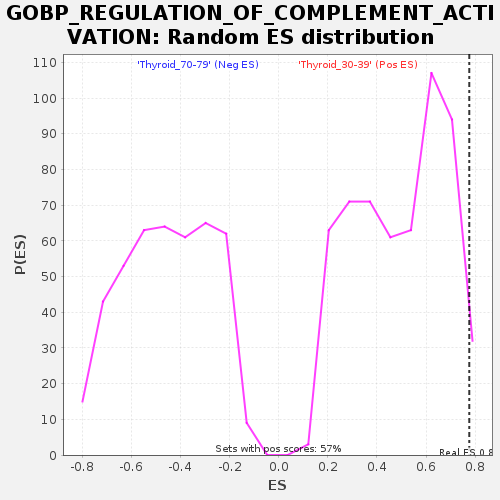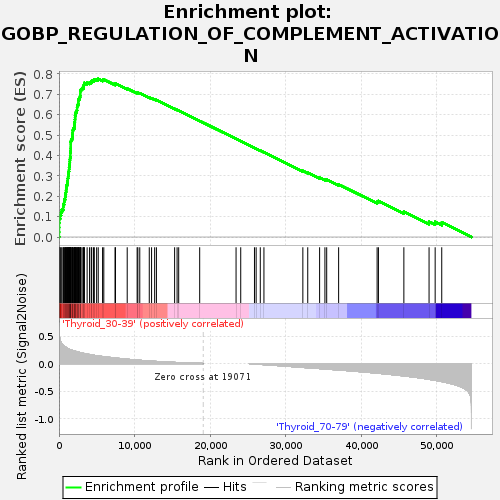
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Thyroid.Thyroid.cls #Thyroid_30-39_versus_Thyroid_70-79.Thyroid.cls #Thyroid_30-39_versus_Thyroid_70-79_repos |
| Phenotype | Thyroid.cls#Thyroid_30-39_versus_Thyroid_70-79_repos |
| Upregulated in class | Thyroid_30-39 |
| GeneSet | GOBP_REGULATION_OF_COMPLEMENT_ACTIVATION |
| Enrichment Score (ES) | 0.7757918 |
| Normalized Enrichment Score (NES) | 1.5671437 |
| Nominal p-value | 0.028318584 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.896 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | C1QC | NA | 22 | 0.551 | 0.0233 | Yes |
| 2 | C1QA | NA | 28 | 0.541 | 0.0465 | Yes |
| 3 | C1QB | NA | 33 | 0.534 | 0.0694 | Yes |
| 4 | C3AR1 | NA | 46 | 0.512 | 0.0912 | Yes |
| 5 | C5AR2 | NA | 167 | 0.419 | 0.1070 | Yes |
| 6 | IGHV2-70 | NA | 266 | 0.384 | 0.1218 | Yes |
| 7 | VSIG4 | NA | 383 | 0.359 | 0.1351 | Yes |
| 8 | CFP | NA | 562 | 0.330 | 0.1460 | Yes |
| 9 | IGKV4-1 | NA | 570 | 0.329 | 0.1600 | Yes |
| 10 | IGHG3 | NA | 685 | 0.317 | 0.1716 | Yes |
| 11 | IGHV3-11 | NA | 702 | 0.315 | 0.1848 | Yes |
| 12 | IGLV2-23 | NA | 825 | 0.303 | 0.1956 | Yes |
| 13 | IGHG2 | NA | 846 | 0.301 | 0.2082 | Yes |
| 14 | C4BPB | NA | 906 | 0.294 | 0.2197 | Yes |
| 15 | IGKV3-15 | NA | 938 | 0.291 | 0.2317 | Yes |
| 16 | IGLV3-25 | NA | 962 | 0.289 | 0.2437 | Yes |
| 17 | IGKV2-40 | NA | 1015 | 0.284 | 0.2550 | Yes |
| 18 | IGKV2-30 | NA | 1114 | 0.276 | 0.2650 | Yes |
| 19 | IGHG1 | NA | 1146 | 0.273 | 0.2762 | Yes |
| 20 | IGKV1-16 | NA | 1158 | 0.272 | 0.2877 | Yes |
| 21 | IGHV3-30 | NA | 1213 | 0.269 | 0.2983 | Yes |
| 22 | IGKV1-5 | NA | 1241 | 0.267 | 0.3093 | Yes |
| 23 | CFHR4 | NA | 1268 | 0.265 | 0.3202 | Yes |
| 24 | IGLV1-47 | NA | 1327 | 0.261 | 0.3304 | Yes |
| 25 | IGKV1D-33 | NA | 1341 | 0.260 | 0.3414 | Yes |
| 26 | IGHV1OR15-1 | NA | 1378 | 0.258 | 0.3518 | Yes |
| 27 | IGLV3-21 | NA | 1420 | 0.255 | 0.3620 | Yes |
| 28 | IGLV3-1 | NA | 1435 | 0.255 | 0.3727 | Yes |
| 29 | IGHV4-59 | NA | 1451 | 0.254 | 0.3834 | Yes |
| 30 | IGHV3-48 | NA | 1478 | 0.252 | 0.3937 | Yes |
| 31 | IGHV3-23 | NA | 1510 | 0.251 | 0.4039 | Yes |
| 32 | IGKV1-12 | NA | 1511 | 0.251 | 0.4147 | Yes |
| 33 | IGHV2-5 | NA | 1536 | 0.249 | 0.4250 | Yes |
| 34 | C2 | NA | 1543 | 0.249 | 0.4356 | Yes |
| 35 | IGHV3-7 | NA | 1550 | 0.248 | 0.4462 | Yes |
| 36 | IGLV7-43 | NA | 1552 | 0.248 | 0.4568 | Yes |
| 37 | IGKV3-20 | NA | 1555 | 0.248 | 0.4675 | Yes |
| 38 | IGLC3 | NA | 1602 | 0.246 | 0.4772 | Yes |
| 39 | IGLV2-8 | NA | 1729 | 0.239 | 0.4852 | Yes |
| 40 | IGKV2D-30 | NA | 1768 | 0.238 | 0.4947 | Yes |
| 41 | IGKV1D-39 | NA | 1794 | 0.236 | 0.5044 | Yes |
| 42 | IGLV2-11 | NA | 1811 | 0.236 | 0.5142 | Yes |
| 43 | IGHV4-34 | NA | 1818 | 0.235 | 0.5242 | Yes |
| 44 | IGHG4 | NA | 1903 | 0.232 | 0.5327 | Yes |
| 45 | IGLV2-14 | NA | 2033 | 0.227 | 0.5401 | Yes |
| 46 | IGLV1-51 | NA | 2044 | 0.227 | 0.5496 | Yes |
| 47 | IGKC | NA | 2057 | 0.226 | 0.5592 | Yes |
| 48 | IGKV2D-28 | NA | 2099 | 0.225 | 0.5681 | Yes |
| 49 | IGHV3-13 | NA | 2110 | 0.224 | 0.5775 | Yes |
| 50 | IGKV1-39 | NA | 2126 | 0.224 | 0.5869 | Yes |
| 51 | IGLV3-19 | NA | 2132 | 0.224 | 0.5964 | Yes |
| 52 | IGLV3-27 | NA | 2209 | 0.221 | 0.6045 | Yes |
| 53 | IGKV5-2 | NA | 2231 | 0.220 | 0.6136 | Yes |
| 54 | CD5L | NA | 2335 | 0.215 | 0.6209 | Yes |
| 55 | IGKV3D-20 | NA | 2398 | 0.213 | 0.6290 | Yes |
| 56 | IGLV1-40 | NA | 2400 | 0.213 | 0.6381 | Yes |
| 57 | IGHV3-33 | NA | 2419 | 0.212 | 0.6469 | Yes |
| 58 | IGLC2 | NA | 2547 | 0.208 | 0.6535 | Yes |
| 59 | IGKV3D-11 | NA | 2573 | 0.207 | 0.6620 | Yes |
| 60 | IGLC6 | NA | 2574 | 0.207 | 0.6709 | Yes |
| 61 | IGHV1-69 | NA | 2627 | 0.205 | 0.6787 | Yes |
| 62 | IGKV2-28 | NA | 2754 | 0.201 | 0.6850 | Yes |
| 63 | SERPING1 | NA | 2784 | 0.199 | 0.6931 | Yes |
| 64 | IGKV1-17 | NA | 2814 | 0.199 | 0.7011 | Yes |
| 65 | IGHV4-39 | NA | 2816 | 0.199 | 0.7096 | Yes |
| 66 | IGKV1D-12 | NA | 2817 | 0.199 | 0.7182 | Yes |
| 67 | IGLC1 | NA | 2899 | 0.195 | 0.7251 | Yes |
| 68 | A2M | NA | 3063 | 0.190 | 0.7303 | Yes |
| 69 | C9 | NA | 3242 | 0.185 | 0.7349 | Yes |
| 70 | CR2 | NA | 3270 | 0.184 | 0.7424 | Yes |
| 71 | IGLV1-44 | NA | 3311 | 0.183 | 0.7495 | Yes |
| 72 | PROS1 | NA | 3365 | 0.182 | 0.7564 | Yes |
| 73 | IGHV3-53 | NA | 3723 | 0.172 | 0.7572 | Yes |
| 74 | CFHR1 | NA | 4045 | 0.163 | 0.7583 | Yes |
| 75 | IGLV6-57 | NA | 4269 | 0.158 | 0.7610 | Yes |
| 76 | C5 | NA | 4330 | 0.156 | 0.7666 | Yes |
| 77 | IGKV2-29 | NA | 4573 | 0.151 | 0.7687 | Yes |
| 78 | C4BPA | NA | 4672 | 0.149 | 0.7732 | Yes |
| 79 | CPB2 | NA | 4984 | 0.143 | 0.7737 | Yes |
| 80 | CR1 | NA | 5194 | 0.139 | 0.7758 | Yes |
| 81 | C1QBP | NA | 5767 | 0.127 | 0.7708 | No |
| 82 | C1S | NA | 5931 | 0.124 | 0.7731 | No |
| 83 | IGLC7 | NA | 7419 | 0.100 | 0.7501 | No |
| 84 | CD19 | NA | 7476 | 0.099 | 0.7534 | No |
| 85 | C4A | NA | 9029 | 0.078 | 0.7282 | No |
| 86 | C1R | NA | 10343 | 0.062 | 0.7068 | No |
| 87 | CD59 | NA | 10439 | 0.061 | 0.7077 | No |
| 88 | C6 | NA | 10692 | 0.058 | 0.7056 | No |
| 89 | CFH | NA | 11950 | 0.046 | 0.6845 | No |
| 90 | C4B | NA | 12255 | 0.043 | 0.6807 | No |
| 91 | CFHR5 | NA | 12663 | 0.039 | 0.6749 | No |
| 92 | C8A | NA | 12895 | 0.038 | 0.6723 | No |
| 93 | C8B | NA | 15305 | 0.020 | 0.6290 | No |
| 94 | F2 | NA | 15633 | 0.018 | 0.6237 | No |
| 95 | CFHR2 | NA | 15845 | 0.017 | 0.6206 | No |
| 96 | CPN1 | NA | 18624 | 0.004 | 0.5697 | No |
| 97 | MIR520E | NA | 23439 | 0.000 | 0.4814 | No |
| 98 | MIR520B | NA | 24057 | 0.000 | 0.4701 | No |
| 99 | C7 | NA | 25876 | -0.007 | 0.4370 | No |
| 100 | C3 | NA | 26097 | -0.010 | 0.4334 | No |
| 101 | PHB | NA | 26646 | -0.015 | 0.4240 | No |
| 102 | CPN2 | NA | 27129 | -0.019 | 0.4160 | No |
| 103 | CR1L | NA | 32281 | -0.069 | 0.3244 | No |
| 104 | CD55 | NA | 32929 | -0.076 | 0.3158 | No |
| 105 | VTN | NA | 34499 | -0.091 | 0.2909 | No |
| 106 | CLU | NA | 35202 | -0.098 | 0.2822 | No |
| 107 | IL1B | NA | 35437 | -0.100 | 0.2823 | No |
| 108 | C5AR1 | NA | 37017 | -0.116 | 0.2583 | No |
| 109 | CFB | NA | 42111 | -0.175 | 0.1723 | No |
| 110 | CFI | NA | 42287 | -0.177 | 0.1767 | No |
| 111 | SUSD4 | NA | 45653 | -0.225 | 0.1246 | No |
| 112 | C8G | NA | 48988 | -0.286 | 0.0757 | No |
| 113 | CD81 | NA | 49796 | -0.305 | 0.0740 | No |
| 114 | CD46 | NA | 50667 | -0.327 | 0.0720 | No |