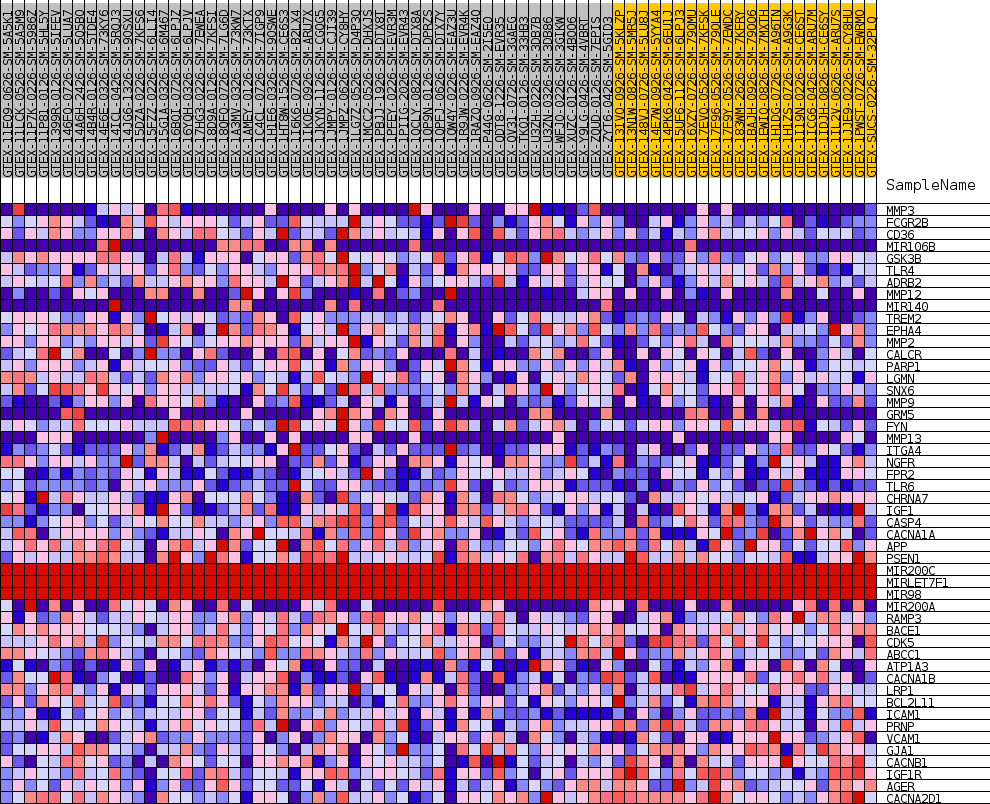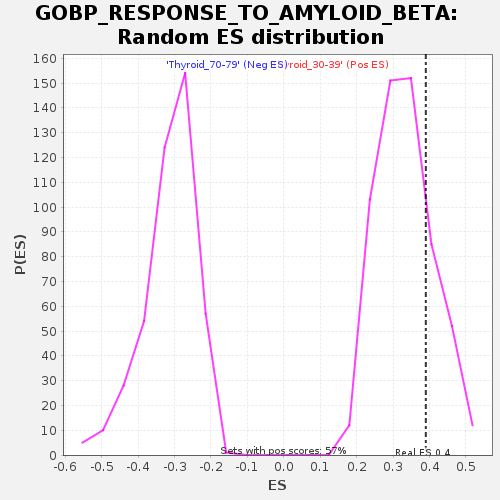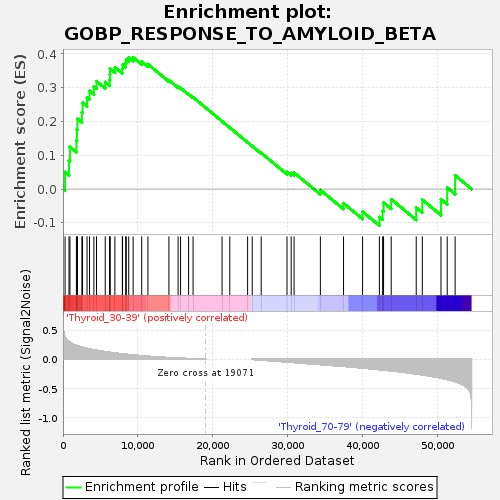
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Thyroid.Thyroid.cls #Thyroid_30-39_versus_Thyroid_70-79.Thyroid.cls #Thyroid_30-39_versus_Thyroid_70-79_repos |
| Phenotype | Thyroid.cls#Thyroid_30-39_versus_Thyroid_70-79_repos |
| Upregulated in class | Thyroid_30-39 |
| GeneSet | GOBP_RESPONSE_TO_AMYLOID_BETA |
| Enrichment Score (ES) | 0.38985988 |
| Normalized Enrichment Score (NES) | 1.1735318 |
| Nominal p-value | 0.21869488 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | MMP3 | NA | 276 | 0.383 | 0.0504 | Yes |
| 2 | FCGR2B | NA | 799 | 0.305 | 0.0850 | Yes |
| 3 | CD36 | NA | 926 | 0.292 | 0.1250 | Yes |
| 4 | MIR106B | NA | 1789 | 0.237 | 0.1435 | Yes |
| 5 | GSK3B | NA | 1859 | 0.234 | 0.1760 | Yes |
| 6 | TLR4 | NA | 1930 | 0.231 | 0.2082 | Yes |
| 7 | ADRB2 | NA | 2524 | 0.209 | 0.2275 | Yes |
| 8 | MMP12 | NA | 2613 | 0.205 | 0.2556 | Yes |
| 9 | MIR140 | NA | 3211 | 0.186 | 0.2716 | Yes |
| 10 | TREM2 | NA | 3542 | 0.176 | 0.2910 | Yes |
| 11 | EPHA4 | NA | 4128 | 0.161 | 0.3037 | Yes |
| 12 | MMP2 | NA | 4477 | 0.153 | 0.3194 | Yes |
| 13 | CALCR | NA | 5640 | 0.130 | 0.3169 | Yes |
| 14 | PARP1 | NA | 6241 | 0.119 | 0.3231 | Yes |
| 15 | LGMN | NA | 6266 | 0.119 | 0.3398 | Yes |
| 16 | SNX6 | NA | 6282 | 0.118 | 0.3567 | Yes |
| 17 | MMP9 | NA | 6933 | 0.108 | 0.3603 | Yes |
| 18 | GRM5 | NA | 7929 | 0.093 | 0.3555 | Yes |
| 19 | FYN | NA | 7957 | 0.092 | 0.3684 | Yes |
| 20 | MMP13 | NA | 8377 | 0.086 | 0.3732 | Yes |
| 21 | ITGA4 | NA | 8453 | 0.085 | 0.3842 | Yes |
| 22 | NGFR | NA | 8780 | 0.081 | 0.3899 | Yes |
| 23 | FPR2 | NA | 9369 | 0.074 | 0.3897 | No |
| 24 | TLR6 | NA | 10518 | 0.060 | 0.3774 | No |
| 25 | CHRNA7 | NA | 11346 | 0.052 | 0.3697 | No |
| 26 | IGF1 | NA | 14153 | 0.028 | 0.3223 | No |
| 27 | CASP4 | NA | 15378 | 0.020 | 0.3027 | No |
| 28 | CACNA1A | NA | 15706 | 0.018 | 0.2992 | No |
| 29 | APP | NA | 16779 | 0.012 | 0.2813 | No |
| 30 | PSEN1 | NA | 17379 | 0.009 | 0.2716 | No |
| 31 | MIR200C | NA | 21252 | 0.000 | 0.2006 | No |
| 32 | MIRLET7F1 | NA | 22285 | 0.000 | 0.1817 | No |
| 33 | MIR98 | NA | 24667 | 0.000 | 0.1381 | No |
| 34 | MIR200A | NA | 25282 | -0.001 | 0.1269 | No |
| 35 | RAMP3 | NA | 26481 | -0.013 | 0.1069 | No |
| 36 | BACE1 | NA | 29929 | -0.046 | 0.0504 | No |
| 37 | CDK5 | NA | 30490 | -0.052 | 0.0476 | No |
| 38 | ABCC1 | NA | 30876 | -0.055 | 0.0485 | No |
| 39 | ATP1A3 | NA | 34388 | -0.090 | -0.0029 | No |
| 40 | CACNA1B | NA | 37483 | -0.121 | -0.0421 | No |
| 41 | LRP1 | NA | 40027 | -0.149 | -0.0671 | No |
| 42 | BCL2L11 | NA | 42272 | -0.177 | -0.0827 | No |
| 43 | ICAM1 | NA | 42705 | -0.182 | -0.0643 | No |
| 44 | PRNP | NA | 42831 | -0.184 | -0.0400 | No |
| 45 | VCAM1 | NA | 43852 | -0.197 | -0.0301 | No |
| 46 | GJA1 | NA | 47196 | -0.251 | -0.0551 | No |
| 47 | CACNB1 | NA | 48009 | -0.266 | -0.0315 | No |
| 48 | IGF1R | NA | 50501 | -0.322 | -0.0306 | No |
| 49 | AGER | NA | 51337 | -0.346 | 0.0043 | No |
| 50 | CACNA2D1 | NA | 52393 | -0.382 | 0.0403 | No |