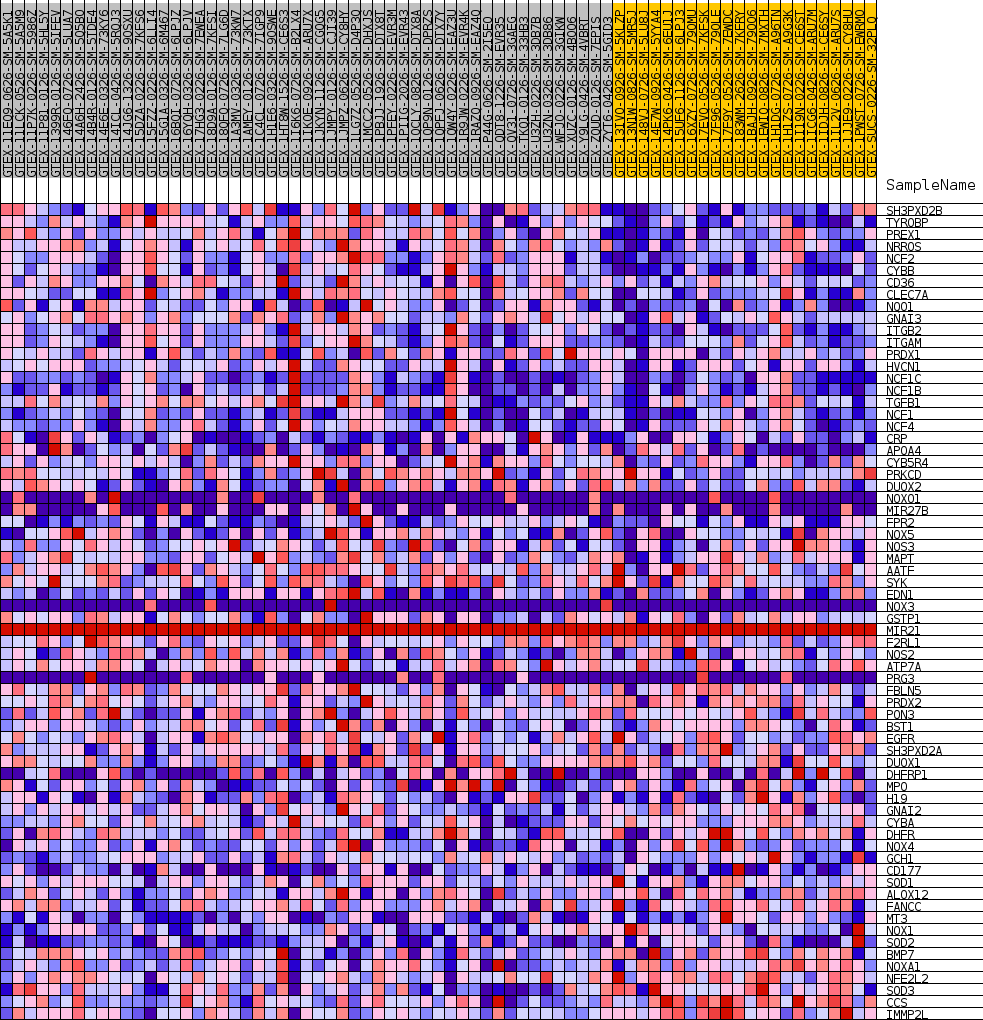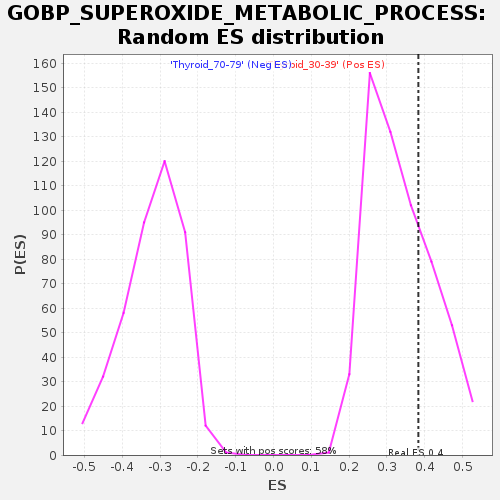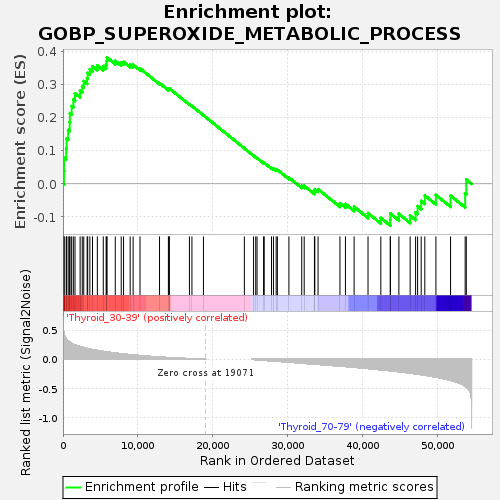
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | Thyroid.Thyroid.cls #Thyroid_30-39_versus_Thyroid_70-79.Thyroid.cls #Thyroid_30-39_versus_Thyroid_70-79_repos |
| Phenotype | Thyroid.cls#Thyroid_30-39_versus_Thyroid_70-79_repos |
| Upregulated in class | Thyroid_30-39 |
| GeneSet | GOBP_SUPEROXIDE_METABOLIC_PROCESS |
| Enrichment Score (ES) | 0.3829921 |
| Normalized Enrichment Score (NES) | 1.142395 |
| Nominal p-value | 0.28892735 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | TITLE | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|---|
| 1 | SH3PXD2B | NA | 127 | 0.442 | 0.0391 | Yes |
| 2 | TYROBP | NA | 152 | 0.425 | 0.0784 | Yes |
| 3 | PREX1 | NA | 427 | 0.351 | 0.1063 | Yes |
| 4 | NRROS | NA | 483 | 0.343 | 0.1373 | Yes |
| 5 | NCF2 | NA | 693 | 0.316 | 0.1631 | Yes |
| 6 | CYBB | NA | 892 | 0.296 | 0.1871 | Yes |
| 7 | CD36 | NA | 926 | 0.292 | 0.2139 | Yes |
| 8 | CLEC7A | NA | 1154 | 0.273 | 0.2352 | Yes |
| 9 | NQO1 | NA | 1382 | 0.258 | 0.2552 | Yes |
| 10 | GNAI3 | NA | 1603 | 0.246 | 0.2741 | Yes |
| 11 | ITGB2 | NA | 2287 | 0.217 | 0.2819 | Yes |
| 12 | ITGAM | NA | 2572 | 0.207 | 0.2961 | Yes |
| 13 | PRDX1 | NA | 2774 | 0.200 | 0.3111 | Yes |
| 14 | HVCN1 | NA | 3227 | 0.185 | 0.3202 | Yes |
| 15 | NCF1C | NA | 3300 | 0.183 | 0.3360 | Yes |
| 16 | NCF1B | NA | 3589 | 0.175 | 0.3471 | Yes |
| 17 | TGFB1 | NA | 3942 | 0.166 | 0.3562 | Yes |
| 18 | NCF1 | NA | 4588 | 0.151 | 0.3585 | Yes |
| 19 | NCF4 | NA | 5382 | 0.135 | 0.3565 | Yes |
| 20 | CRP | NA | 5763 | 0.127 | 0.3615 | Yes |
| 21 | APOA4 | NA | 5848 | 0.126 | 0.3717 | Yes |
| 22 | CYB5R4 | NA | 5871 | 0.125 | 0.3830 | Yes |
| 23 | PRKCD | NA | 6985 | 0.107 | 0.3726 | No |
| 24 | DUOX2 | NA | 7779 | 0.095 | 0.3669 | No |
| 25 | NOXO1 | NA | 8090 | 0.091 | 0.3697 | No |
| 26 | MIR27B | NA | 8975 | 0.078 | 0.3608 | No |
| 27 | FPR2 | NA | 9369 | 0.074 | 0.3605 | No |
| 28 | NOX5 | NA | 10288 | 0.063 | 0.3496 | No |
| 29 | NOS3 | NA | 12898 | 0.038 | 0.3052 | No |
| 30 | MAPT | NA | 14082 | 0.028 | 0.2862 | No |
| 31 | AATF | NA | 14176 | 0.028 | 0.2871 | No |
| 32 | SYK | NA | 14202 | 0.028 | 0.2892 | No |
| 33 | EDN1 | NA | 16892 | 0.011 | 0.2409 | No |
| 34 | NOX3 | NA | 17228 | 0.010 | 0.2357 | No |
| 35 | GSTP1 | NA | 18764 | 0.003 | 0.2078 | No |
| 36 | MIR21 | NA | 24232 | 0.000 | 0.1075 | No |
| 37 | F2RL1 | NA | 25460 | -0.003 | 0.0853 | No |
| 38 | NOS2 | NA | 25752 | -0.006 | 0.0805 | No |
| 39 | ATP7A | NA | 25927 | -0.008 | 0.0781 | No |
| 40 | PRG3 | NA | 26797 | -0.016 | 0.0637 | No |
| 41 | FBLN5 | NA | 26884 | -0.017 | 0.0637 | No |
| 42 | PRDX2 | NA | 27857 | -0.026 | 0.0483 | No |
| 43 | PON3 | NA | 28119 | -0.028 | 0.0462 | No |
| 44 | BST1 | NA | 28479 | -0.032 | 0.0426 | No |
| 45 | EGFR | NA | 28643 | -0.033 | 0.0427 | No |
| 46 | SH3PXD2A | NA | 30183 | -0.049 | 0.0190 | No |
| 47 | DUOX1 | NA | 31897 | -0.066 | -0.0062 | No |
| 48 | DHFRP1 | NA | 32228 | -0.069 | -0.0058 | No |
| 49 | MPO | NA | 33594 | -0.082 | -0.0232 | No |
| 50 | H19 | NA | 33644 | -0.083 | -0.0164 | No |
| 51 | GNAI2 | NA | 34078 | -0.087 | -0.0162 | No |
| 52 | CYBA | NA | 37003 | -0.116 | -0.0590 | No |
| 53 | DHFR | NA | 37738 | -0.123 | -0.0609 | No |
| 54 | NOX4 | NA | 38898 | -0.136 | -0.0695 | No |
| 55 | GCH1 | NA | 40755 | -0.157 | -0.0888 | No |
| 56 | CD177 | NA | 42467 | -0.179 | -0.1034 | No |
| 57 | SOD1 | NA | 43728 | -0.196 | -0.1082 | No |
| 58 | ALOX12 | NA | 43746 | -0.196 | -0.0901 | No |
| 59 | FANCC | NA | 44879 | -0.212 | -0.0910 | No |
| 60 | MT3 | NA | 46382 | -0.236 | -0.0964 | No |
| 61 | NOX1 | NA | 47111 | -0.249 | -0.0865 | No |
| 62 | SOD2 | NA | 47381 | -0.253 | -0.0677 | No |
| 63 | BMP7 | NA | 47861 | -0.263 | -0.0519 | No |
| 64 | NOXA1 | NA | 48343 | -0.272 | -0.0352 | No |
| 65 | NFE2L2 | NA | 49827 | -0.305 | -0.0338 | No |
| 66 | SOD3 | NA | 51785 | -0.361 | -0.0359 | No |
| 67 | CCS | NA | 53734 | -0.460 | -0.0286 | No |
| 68 | IMMP2L | NA | 53888 | -0.474 | 0.0129 | No |